Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
HEMATOLOGY / RESEARCH PAPER
Comprehensive Expression of Long non-coding RNAs and association with the Iron and Erythropoiesis Regulatory Proteins in Transfusion-dependent β-Thalassemia
1
Department of Medical Laboratory Sciences, Faculty of Allied Medical Sciences, Al-Balqa Applied University, Jordan
2
Key Laboratory of Laboratory Medicine, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Zhejiang, China
3
Arkan Laboratory, Zarqa, Jordan
4
Department of Allied Medical Sciences, Al-Balqa Applied University, Jordan
5
Department of Medical Laboratory Sciences, Faculty of Allied Medical Sciences, Al-Ahliyya University, Amman, Jordan
6
Department of Biology, School of Science, Jordan University, Amman, Jordan
7
Biolab Diagnostic Laboratories, Amman, Jordan
Submission date: 2025-02-12
Final revision date: 2025-05-09
Acceptance date: 2025-06-01
Online publication date: 2025-06-22
Corresponding author
Ola M. Al-Sanabra
Department of Medical Laboratory Sciences, Faculty of Allied Medical Sciences, Al-Balqa Applied University, 19117, Al-Salt, Jordan
Department of Medical Laboratory Sciences, Faculty of Allied Medical Sciences, Al-Balqa Applied University, 19117, Al-Salt, Jordan
KEYWORDS
TOPICS
ABSTRACT
Introduction:
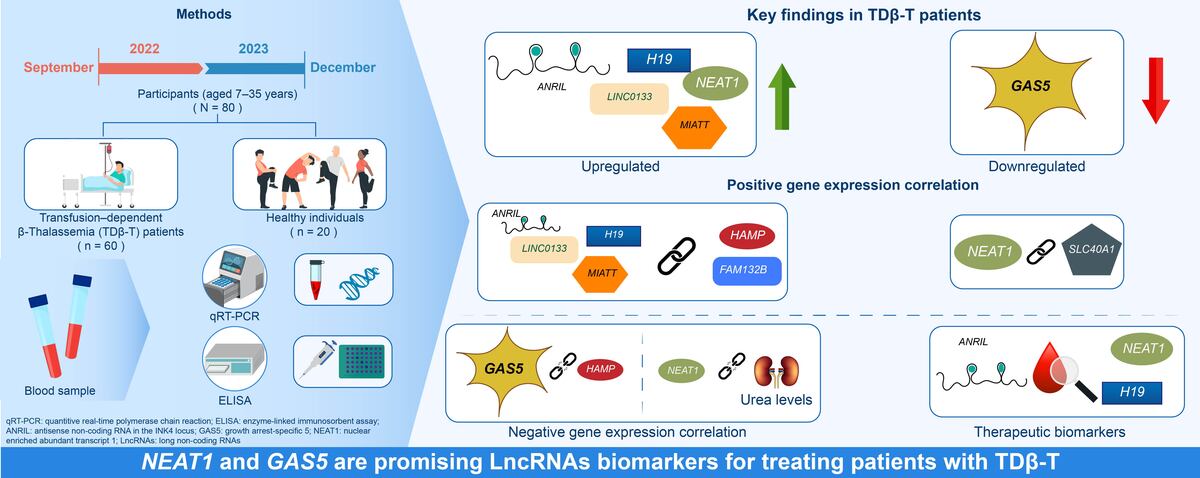
β-Thalassemia is a genetic disorder characterized by a quantitative defect in β-globin synthesis caused by genetic and epigenetic alterations. However, the expression patterns of long non-coding RNAs (LncRNAs) and their relationship with genes and proteins involved in iron metabolism and erythropoiesis remain largely unknown. We aimed to investigate the expression of LncRNAs and their correlation with iron and erythropoiesis regulatory proteins in patients with transfusion dependent-β-Thalassemia (TDβ-T).
Material and methods:
Whole blood samples and clinical records were collected from 60 patients with TDβ-T and 20 healthy controls. Expression levels of selected LncRNAs were measured using qRT-PCR. Iron metabolism and erythropoiesis-related proteins were quantified using ELISA.
Results:
TDβ-T patients exhibited significantly elevated levels of iron and erythropoiesis-regulating proteins, as well as increased expression of HAMP, GDF-15, FAM132B, and SLC40A1 compared to controls. Additionally, LncRNAs ANRIL, H9, LINCO133, MIAT, and NEAT1 were markedly upregulated, while LncRNA GAS5 was downregulated in patients with TDβ-T. Among these, LncRNAs NEAT1 and GAS5 showed the strongest diagnostic performance. A significant correlation was observed between the expression of HAMP and FAM132B and LncRNAs ANRIL, H19, LINCO133, and MIAT. Furthermore, LncRNA NEAT1 expression correlated positively with SLC40A1 and negatively with urea levels, whereas LncRNA GAS5 was inversely correlated with HAMP expression.
Conclusions:
This study is the first to demonstrate altered LncRNA expression patterns and their associations with iron metabolism, erythropoiesis-regulating proteins, and urea levels in patients with TDβ-T. These findings provide new insights for future research and potential therapeutic targets.
β-Thalassemia is a genetic disorder characterized by a quantitative defect in β-globin synthesis caused by genetic and epigenetic alterations. However, the expression patterns of long non-coding RNAs (LncRNAs) and their relationship with genes and proteins involved in iron metabolism and erythropoiesis remain largely unknown. We aimed to investigate the expression of LncRNAs and their correlation with iron and erythropoiesis regulatory proteins in patients with transfusion dependent-β-Thalassemia (TDβ-T).
Material and methods:
Whole blood samples and clinical records were collected from 60 patients with TDβ-T and 20 healthy controls. Expression levels of selected LncRNAs were measured using qRT-PCR. Iron metabolism and erythropoiesis-related proteins were quantified using ELISA.
Results:
TDβ-T patients exhibited significantly elevated levels of iron and erythropoiesis-regulating proteins, as well as increased expression of HAMP, GDF-15, FAM132B, and SLC40A1 compared to controls. Additionally, LncRNAs ANRIL, H9, LINCO133, MIAT, and NEAT1 were markedly upregulated, while LncRNA GAS5 was downregulated in patients with TDβ-T. Among these, LncRNAs NEAT1 and GAS5 showed the strongest diagnostic performance. A significant correlation was observed between the expression of HAMP and FAM132B and LncRNAs ANRIL, H19, LINCO133, and MIAT. Furthermore, LncRNA NEAT1 expression correlated positively with SLC40A1 and negatively with urea levels, whereas LncRNA GAS5 was inversely correlated with HAMP expression.
Conclusions:
This study is the first to demonstrate altered LncRNA expression patterns and their associations with iron metabolism, erythropoiesis-regulating proteins, and urea levels in patients with TDβ-T. These findings provide new insights for future research and potential therapeutic targets.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.