Introduction
Pancreatic cancer as a highly malignant tumor is predicted to be the second cause of cancer-associated death in the next decade [1, 2]. Rapid spread in patients and difficulty of detection in the early stages have made pancreatic cancer become the leading cause of cancer-related death [3]. The cancer mortality is extremely high, though substantial improvement has been achieved in the fields of early diagnosis, surgical techniques, combination chemotherapies and other anti-cancer treatments for patients with pancreatic cancer [4, 5]. There are no apparent symptoms or signs in patients unless this malignancy progresses to an advanced stage [6]. Once in advanced stages, the chance of surgery has been lost. Therefore, it is of vital importance that clinical physicians should identify the patients at high risk. The cancer-specific 5-year overall survival is less than 5% [7]. Moreover, patients who have undergone curative surgical resection still relapse in the following 2 years. Hence, the mechanisms of cancer development and progression need to be revealed and cancer treatments, potential biomarkers and novel anti-cancer strategies urgently need to be identified [8].
Integrins – adhesion receptors that play an important role in signaling from the extracellular matrix to the cell – are composed of an α chain and a β chain [9, 10]. Integrins mediate critical cell signaling through acting as key elements of conducting inside-out and outside-in signaling. For heterodimeric trans-membrane proteins, the subunits are capable of forming different heterodimers [11, 12]. The integrin subunit β 6 (ITGB6) gene encodes a protein that belongs to the integrin superfamily and as a vital subunit of the integrin αvβ6 heterodimer can regulate this heterodimer protein’s expression level in different status [13, 14]. The integrin αvβ6 as a receptor of ECM proteins and TGF-β’s latency-associated peptide regulates key biological processes, including fetal development and wound healing [15–17]. Previous studies also reported that it might be involved in pathological processes and carcinogenesis [18, 19]. The integrin αvβ6 is constitutively expressed in healthy epithelia, normally, while during carcinogenesis the expression level is up-regulated [20]. Moreover, ITGB6 mainly regulates αvβ6 expression owing to its important role in forming a heterodimer with αv proteins. ITGB6 is largely expressed in developing tissues, wound healing and human epithelial malignancies but not in the normal epithelia. Recent studies showed that ITGB6 plays an important role in multiple human cancers, such as brain cancer, colon cancer and oral cancer, and is related to the clinical progression [21–23]. Hence, it is of vital importance to uncover the ITGB6 role in pancreatic cancer.
The present study demonstrated that high ITGB6 expression was significantly correlated with poor clinical outcomes in pancreatic cancer patients and further functional assays revealed that knockdown ITGB6 in vivo and in vitro might inhibit cancer cell proliferation and the ability of invasion or migration. According to the data we present, ITGB6 may be a prognostic biomarker for pancreatic cancer and serve as a novel target in the clinical treatment.
Material and methods
In this section, we followed the methods of Zheng et al. 2015 and Niu et al. 2017 [24, 25].
Patient and public involvement
This retrospective study identified 82 consecutive patients with PDAC who underwent pancreatic cancer resection in our hospital between May 2012 and May 2017. Patients were classified according to the 2009 UICC TNM staging and the 2004 WHO/ISUP classification. All patients had no preoperative radiotherapy or chemotherapy and no neoadjuvant chemotherapy. Participants provided written informed consent. All patients showed no evidence of tumor metastasis confirmed by cross-sectional imaging.
Immunohistochemistry
Pancreatic cancer specimens were stained for detection of ITGB6 expression. Pancreatic cancer tissues were cut in sections 3.5 μm thick and pretreated in Tris-buffered saline. After antigen retrieval, samples were incubated with primary antibody rabbit anti-ITGB6 (1 : 200 dilution, ab233519, Abcam, Cambridge, UK) at 4°C overnight. Next, sections were washed twice with Tris-buffered saline and incubated for 30 min at room temperature with the blocking buffer of 5% conjugated secondary antibody. Subsequently, diaminobenzidine (DAB) was dripped on in 3–5 min. The images were collected by microscope (Nikon, Tokyo, Japan).
ITGB6 protein is mainly located in the cell membrane of tumor cells. The tissues were scored according to the proportion of positive tumor cells and the staining intensity of tumor cells. The proportion of positive tumor cells < 10% scored 0; the percentage of positive tumor cells occupying 10% to 30% scored 1. The positive percentage of tumor cells of 30–70% scored 2, and the positive percentage of tumor cells of 70% > scored 3. The positive staining was evaluated zero for negative staining, 1 for weak positive staining, 2 for moderate positive staining, and 3 for strong positive staining. The final scores were calculated by multiplying the proportion and intensity. Results of high expression (> 3) or low expression (0–3) were determined according to the score of the positive cell percentage score and the staining intensity score. The sections of each patient were observed within 3 visual fields, and two experienced pathologists read the sections without knowing the pathological grade and clinical data. The results were evaluated by the double-blind method.
Cell culture and reagents
Human pancreatic cancer cell lines PANC-1 and BxPC-3 were purchased from the China Center for Type Culture Collection. The above cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) and RPMI, respectively, all supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 0.1 mg/ml streptomycin. All cells were maintained at 37°C in a 5% CO2 humidified incubator.
Stable cell line generation
The human ITGB6 (gene ID: 80381) targeting small hairpin (sh) RNA was purchased from Genechem (Lot: TR312078, Genechem Co., Ltd, Shanghai, China) and non-effective 29-mer scrambled shRNA (Lot: TR30012; Genechem Co., Ltd, Shanghai, China). Recombinant lentivirus was titered to 2 × 108 TU/ml, and cells of interest were transfected by the lentivirus with the multiplicity of infection (MOI) of 20. RT-qPCR and Western blotting were performed to confirm the effect of ITGB6 silencing.
RNA analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) and the RNA integrity confirmed. A PrimeScript RT reagent kit (Invitrogen) was used for cDNA generation. RT-qPCR was performed with SuperReal PreMix (Tiangen Biotech) by the following program: 95°C for 3 min in 1 cycle; 95°C for 5 s, 58°C for 30 s, and 72°C for 30 s in 35 cycles. To ensure the DNA production, a melting curve analysis was performed according to the ABI StepOne system. The relative gene expression was normalized to the internal standard β-actin using the 2-∆∆Ct method. The whole experiment was performed independently three times. Primers used for the analysis were as follows:
ITGB6 F: 5′-CTTTCCCTAGCCTGCCTTCT-3′,
ITGB6 R: 5′-GTTCAATCCCCATCCGTTT-3′,
β-actin F: 5′-CAGCTCACCATGGATGATGATATC-3′,
β-actin R: 5′-AAGCCGGCCTTGCACAT-3′.
Immunoblotting
Cells were lysed in lysis buffer (60 mM Tris-HCl, pH 6.8, 2% SDS, 20% glycerol, 0.25% bromophenol blue, 1.25% 2-mercaptoethanol and protease inhibitor cocktail). Lysates were subjected to the BCA Protein Assay (Thermo Fisher Scientific) to measure the concentration following the manufacturer’s instructions. Next, equal amounts of protein (30–50 mg) were electrophoresed by 10% SDS-PAGE gels and transferred onto PVDF membranes (Merck Millipore). Then, using non-fat milk to block the membrane, the membranes were incubated with antibodies at 4°C overnight. The antibodies used were as follows: rabbit anti-ITGB6 (1 : 1000 dilution, ab233519, Abcam, Cambridge, UK); mouse anti-β-actin (1 : 1000 dilution, ab8226, Abcam, UK), rabbit anti-Ki67 (1 : 1000 dilution, ab16667, Abcam, UK), mouse anti-proliferating cell nuclear antigen (PCNA) (1 : 500 dilution, ab29, Abcam, UK), mouse anti-MMP2 (1 : 1000 dilution, ab37150, Abcam, UK) and mouse anti-MMP9 (1 : 1000 dilution, ab38898, Abcam, UK).
Colony formation assays
Cell proliferation ability was evaluated by colony formation assay. 400 cells per well were plated in 6-well plates. Cells were fixed in methyl hydrate for 10 min, accordingly, and the colonies were stained and counted using an optical microscope.
MTT assay
MTT assay was used to study cell proliferation ability. 3 × 103 cells were plated in a 96-well plate, and 10 μl of 5 mg/ml MTT was added to each well and left to culture for 4 h. After adding dimethyl sulfoxide to the plate, the plate was placed on a microplate autoreader (Bio-Rad, Hercules) and the optical density at 570 nm wavelength was acquired. Cell growth curves were generated to determine the optical density value.
Cell scratch assays
The cells were seeded in 6-well plates and grew into full confluence overnight. A sterile pipette tip was used to introduce a scratch in the middle of each well. Next, the growth medium was discarded and replaced with fresh medium. The rate of migration towards the center of the wound was ascertained at indicated time points.
Cell invasion assays
In migration or invasion assay, 3 × 105 cells were plated in the upper chambers coated with Matrigel (BD Bioscience, Mountain View, CA, USA). The growth medium was added into the lower chamber and cultured at 37°C for 24 h. Subsequently, cells that migrated to the lower side of membrane were fixed and stained. The cells were calculated at three randomly selected fields by microscope from each chamber.
Growth assay in vivo
The in vivo animal study was approved by Tianjin Medical University Cancer Institute and Hospital. The immunocompromised nude mice (aged 4–5 weeks) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The 1 × 106 non-affected shRNA treated cells and 1 × 106 ITGB6 shRNA treated cells were injected subcutaneously into the mice. After 4 weeks of inoculation, the final volume of tumor was weighed.
Statistical analysis
All data were analyzed with SPSS 22.0 software. The connection between ITGB6 expression level and patients’ survival time after surgery was performed by the Kaplan-Meier method. Statistical comparisons were conducted by Student’s t-test. The categorical data were analyzed by the χ2 method. The value of statistical significant was considered as p < 0.05.
Results
Increased expression of ITGB6 was associated with poor clinical outcomes in pancreatic cancer patients
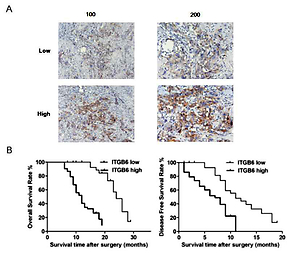
We divided the 78 pancreatic adenocarcinoma patients into two groups – the ITGB6 low group (n = 28) and ITGB6 high group (n = 50) – based on the protein expression level of ITGB6 (Table I). Immunohistochemistry was used for detection of ITGB6 expression with low and high intensity in stained pancreatic cancer specimens (Figure 1 A). Particularly, high ITGB6 protein expression level was strongly associated with high TNM stage (p = 0.040), large tumor size (p = 0.039) and vascular invasion (p = 0.030) when compared to the low expression group. No significant clinical connection was determined between ITGB6 expression and other clinicopathological features, including age, gender, tumor grade, and lymph node metastasis (Table I). To better investigate the ITGB6 clinical features, Kaplan-Meier survival analysis was performed to determine the cancer overall survival (OS) in our 30-month follow-up time after surgery. The data showed that high ITGB6 protein expression level was related to a short cancer OS time and disease-free survival time (both p < 0.05, Figure 1 B).
Figure 1
Increased expression of ITGB6 was associated with poor clinical outcomes in our clinical data. A – Low and high IGTB6 expression in patients by immunohistochemistry is shown. B – Overall survival (OS) and disease-free survival (DFS) rates with ITGB6 expression in our clinical data: high ITGB6 protein expression level was significantly associated with a shorter cancer OS time and DFS time compared with the low expression group (both p < 0.05)
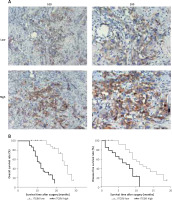
Table I
Relationships of ITGB6 and clinicopathological characteristics in our clinical data of 78 patients with pancreatic adenocarcinoma
To determine the ITGB6 mRNA expression level, bioinformation analysis was performed in the interactive web server GEPIA with the sequencing expression data of 9736 tumors. MRNA expression level of ITGB6 in PAAD (pancreatic adenocarcinoma) was significantly higher than in normal tissues (p < 0.05, PAAD number = 179, normal tissues number = 171, Figure 2 A). We also investigated the cancer-specific survival in the PAAD dataset in GEPIA and found that high ITGB6 expression was significantly associated with shorter OS and disease-free survival in PAAD patients (p = 0.025 and p = 0.041, Figures 2 B, C).
Figure 2
Increased expression of ITGB6 was associated with poor clinical outcomes in TCGA data. A – Expression level of ITGB6 in cancer and normal tissues (PAAD: pancreatic adenocarcinoma; *p < 0.05). B – Overall survival (OS) and disease-free survival (DFS) rates with ITGB6 expression in TCGA pancreatic adenocarcinoma data: high ITGB6 expression group (n = 89) was significantly associated with lower cancer OS rate and disease-free survival rate compared with low expression group (n = 89; p = 0.025 and p = 0.041, respectively)
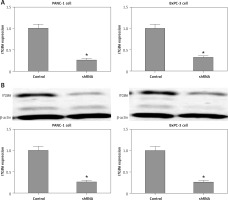
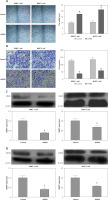
Construction of lentivirus-based ITGB6 stable knockdown cell lines of PANC-1 and BxPC-3
PANC-1 and BxPC-3 cells were transfected with recombinant lentivirus to silence ITGB6 expression in cancer cells. Through RT-qPCR and Western blotting validation, we generated stable cell lines with both ITGB6 mRNA and protein levels significantly lower than in control cells (Figure 3). Using this model, we could efficiently study the role of ITGB6 in cancer cells.
Figure 3
mRNA and protein expression level of ITGB6 in generated stable cell lines. Both mRNA and protein expression levels of ITGB6 were significantly decreased in (A) RT-qPCR and (B) western blotting compared to control cells. All results are obtained from at least three independent experiments (*p < 0.05)
ITGB6 knockdown reduced proliferation through regulating Ki67 and PCNA proteins in PANC-1 and BxPC-3 cell lines
To investigate the biological effect of ITGB6, we conducted the colony formation assay and MTT assay to evaluate the possible role for proliferation in pancreatic cancer cell lines PANC-1 and BxPC-3. Compared to control cells, stable knockdown of ITGB6 in PANC-1 and BxPC-3 cells by recombinant lentivirus significantly decreased the ability of proliferation in vitro compared to control cells (p < 0.05, Figures 4 A, B). The above results may imply that ITGB6 knockdown reduced proliferation ability in cancer cells. To further investigate the mechanism of partially eliminated proliferation, we checked proliferation related proteins such as Ki67 and PCNA in vitro. Western blotting revealed that Ki67 and PCNA proteins were significantly down-regulated in the silenced ITGB6 cell lines compared with the control (p < 0.05, Figures 4 C, D). Taken together, the above data suggested that ITGB6 knockdown may down-regulate Ki67 and PCNA protein to reduce the proliferation in pancreatic cancer cells.
Figure 4
Role of ITGB6 in proliferation in PANC-1 and BxPC-3 cell lines. A – Colony formation assay. Results showed that compared to control cells, stable knockdown ITGB6 in PANC-1 and BxPC-3 cells by recombinant lentivirus significantly decreased the ability of proliferation in vitro (p < 0.05). B – MTT assay. Decreased OD value in ITGB6 silencing group was observed compared to control in vitro (p < 0.05). C – Western blotting revealed that Ki67 and PCNA, proliferation related proteins, were significantly decreased in ITGB6 silencing group compared to control group (both p < 0.05). All results are obtained from at least three independent experiments (*p < 0.05). D – Western blotting revealed that Ki67 and PCNA, proliferation related proteins, were significantly decreased in ITGB6 silencing group compared to control group (both p < 0.05). All results are obtained from at least three independent experiments (*p < 0.05)
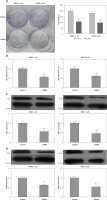
Through down-regulation of MMP2 and MMP9, ITGB6 silencing inhibited migration or invasion in vitro
Cell scratch assay was used to study the cell migration changes. Compared with non-effective scrambled shRNA treated cells, the ITGB6 silenced cells showed a bigger scratch space in PANC-1 and BxPC-3 cell lines (both p < 0.05, Figure 5 A). To investigate the possible role of regulating the cancer cells’ invasive capacity in stable knockdown ITGB6 cells, cell invasion assays were performed. It is obvious that the invasive capacity of ITGB6 shRNA treated cells tended to be partially lost compared to control cells after 48 h (both p < 0.05, Figure 5 B). Moreover, to research the possible mechanism of the above changes, we checked invasion and metastasis associated proteins including matrix metalloproteinase (MMP)-2 and MMP9 at the protein level by immunoblotting. The results showed that MMP2 and MMP9 in ITGB6 shRNA treated cells were lower than the control (both p < 0.05, Figures 5 C, D). The above data suggested that through regulating the associated proteins, knockdown ITGB6 might inhibit the cancer cells’ invasive capacity.
Figure 5
Role of ITGB6 in migration and invasion in PANC-1 and BxPC-3 cell lines. A – Cell scratch assays. Results showed that compared with the control, the ITGB6 silencing cells showed a bigger scratch space in PANC-1 and BxPC-3 cell lines (p < 0.05). B – Cell invasion assays. The results showed that the invasive cell numbers in the ITGB6 shRNA treated group were significantly lower than in the control group (p < 0.05). C, D – Western blotting showed that MMP2 and MMP9, metastasis associated proteins, were significantly decreased in the ITGB6 silencing group compared to the control group (both p < 0.05). All results are from at least three independent experiments (*p < 0.05)
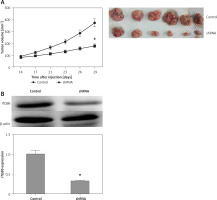
In vivo inhibitory role of ITGB6 knockdown
Based on the results above, down-regulated ITGB6 in pancreatic cancer cell lines significantly inhibited cell proliferation and the ability of invasion or migration in vitro. To better investigate the above findings in vivo, xenograft assay was performed. The tumor volume of the down-regulated ITGB6 group was observably smaller than the control after 4 weeks of treatment (p < 0.05, Figure 6 A). In addition to further confirming the effect of ITGB6 silencing, western blotting assay was performed to consolidate the ITGB6 silencing effect in mouse tumors. The following data showed that ITGB6 was dramatically reduced by our recombinant lentivirus (p < 0.05, Figure 6 B). Taken together, our results suggested that ITGB6 knockdown may exert an inhibitory effect in vivo.
Figure 6
Inhibitory role of ITGB6 silencing in vivo. A – Xenograft assay. After 4 weeks of management, the tumor volume of the down-regulated ITGB6 group was observably smaller than the control group (p < 0.05). B – Western blotting showed that ITGB6 was dramatically reduced by our recombinant lentivirus in injected tumor xenografts in nude mice (p < 0.05). All results are from at least three independent experiments (*p < 0.05)
Discussion
Pancreatic cancer is predicted to be the second cause of cancer-associated death in the next decade [1, 2]. Recent studies have suggested that αvβ6 was overexpressed in various types of tumor and is closely associated with invasive and metastatic features of cancer [23, 26, 27]. It is suggested that ITGB6 mainly regulates αvβ6 expression owing to its vital role in forming a heterodimer with αv proteins. ITGB6 is largely expressed in developing tissues, wound healing and human epithelial malignancies but not in the normal epithelia. Recent studies showed that ITGB6 plays an important role in multiple human cancers, such as brain cancer, colon cancer and oral cancer, and is related to clinical progression [21–23]. Therefore, ITGB6 might play a role in pancreatic cancer progression.
Our present study revealed that ITGB6 was not expressed or had low expression in normal pancreatic tissue compared with tumor, since ITGB6 was not expressed or showed low expression in the normal epithelia pancreatic tissue. Moreover, we performed in-silico TCGA analysis by GEPIA and validated that the mRNA expression level of ITGB6 in PAAD was significantly higher than normal. Further, cancer-specific survival of PAAD was also analyzed and the results showed that high ITGB6 expression was significantly connected with shorter OS and disease-free survival in PAAD patients. Our clinical data showed that patients with high ITGB6 expression in immunohistological analysis are more likely to have a high TNM stage, large tumor size and vascular invasion, and their cancer OS time and disease-free survival time were lower than in the low expression group. Our results showed that the high ITGB6 expression group had a significantly higher percentage of lymph node metastasis, in accordance with various studies concerning the function of integrin β6 in metastasis [28–30].
Several studies have indicated that ITGB6 may promote cancer from the in situ to the invasive stage and from the invasive stage to regional and distant metastasis in different human epithelial malignancies [22, 23, 31]. Krisha et al. found that HER2+ breast cancer patients had elevated levels of ITGB6 and increased regional and distant metastasis, which might be mediated by ITGB6 through the canonical Rho–Rac pathway [28]. To confirm this clinical finding, we generated ITGB6 targeting shRNA with recombinant lentivirus to silence its expression in pancreatic cancer cells, then validated it by western blot assay. We investigated some metastasis-associated proteins expression, such as MMP2 and MMP-9, while silencing ITGB6. Consistent with these results, the mechanism of metastasis suggested that ITGB6 regulated MMP2 and MMP9 to influence the cancer invasive abilities [32]. In breast cancer, Moore et al. demonstrated that use of antibody (264RAD) targeting ITGB6 alone or plus trastuzumab might benefit patients with high-risk and trastuzumab-resistant breast cancer [33]. In immunotherapy, Whilding et al. evaluated targeting αvβ6 chimeric antigen receptor-engineered T cells in which expression of this integrin was aberrant and developed a therapy in phase 1 for patients with other untreatable αvβ6-expressing tumors [34]. In addition, our results showed that ITGB6 knockdown greatly reduced proliferation through regulating Ki67 and PCNA proteins in PANC-1 and BxPC-3 cell lines in vitro. In recent years, many biomarkers have been given more attention for diagnosis and treatment of tumor and non-tumor diseases [35–39]. In summary, the present study demonstrated that silencing ITGB6 in pancreatic cancer cell lines might inhibit the cell behaviors by regulating the proteins involved. However, the key signaling pathway and molecular mechanism of ITGB6 still need to be elucidated.
Strengths and limitations of this study
We first reported that increased expression of ITGB6 was significantly correlated with poor clinical outcome in both our clinical data and TCGA data of pancreatic cancer. Furthermore, functional assays revealed that ITGB6 knockdown in vivo and in vitro might inhibit cancer cell proliferation and the ability of invasion or migration. Our data suggest that ITGB6 is associated with pancreatic cancer malignant progression. Hence, ITGB6 may serve as a potential target of pancreatic cancer in clinical treatment. However, the results should be corroborated by multi-center studies with more patients, and the mechanism of ITGB6 in pancreatic cancer should be further explored.
In conclusion, we firstly demonstrated that elevated ITGB6 expression was significantly correlated with poor clinical outcome in pancreatic cancer. Then we studied the functional role of ITGB6 by knockdown in vitro and in vivo observed significant inhibition of cancer cell proliferation and the ability of invasion or migration. Hence, ITGB6 might be a potential target for the future study of pancreatic cancer.