Introduction
Multiple myeloma (MM) is an incurable hematologic malignancy characterized by plasma cell abnormal proliferation in the bone marrow [1, 2]. Despite great progress in treatment strategies such as chemoradiotherapy and hematopoietic stem cell transplantation, the survival rate of MM patients is still unsatisfactory [3–5]. Therefore, it is urgent to find new diagnostic markers and potential therapeutic targets for MM.
Long noncoding RNA (lncRNA) is an RNA molecule that is widely found in various organisms and has a transcript length of more than 200 nt [6]. Numerous studies have shown that lncRNAs are involved in human disease development, including MM [7–9]. For example, lncRNA AL928768.3 was found to be overexpressed in MM patients and its silencing could repress MM cell proliferation [10]. Also, downregulation of lncRNA BDNF-AS has been confirmed to hinder MM tumor growth [11]. Nuclear enriched abundant transcript 1 (NEAT1) is a key lncRNA that plays a vital role in tumor cell growth [12, 13]. Previous studies have shown that high expression of NEAT1 in MM patients was not only associated with chemoresistance, but also might predict poor prognosis [14, 15]. Therefore, NEAT1 may be an important regulator of the MM process, and its underlying molecular mechanism is still worth further investigation.
LncRNA can act as a competitive endogenous RNA to serve as a molecular sponge for microRNA (miRNA), and then indirectly regulate mRNA expression [16, 17]. MiR-133a, a tumor suppressor, participates in the regulation of various cancer processes, such as colorectal cancer [18] and esophageal cancer [19]. Nevertheless, the role of miR-133a in MM needs to be further explored. Actin-related protein 2/3 complex subunit 5 (ARPC5) belongs to the ARP2/3 complex family and mainly mediates actin assembly [20]. In an MM-related study, ARPC5 was found to be overexpressed and might be used as a biomarker to predict poor prognosis for MM patients [21]. However, whether ARPC5 mediates the malignant progression of MM remains unclear.
The aim of this study was to reveal the underlying molecular mechanism of NEAT1 regulating MM progression. Through bioinformatics analysis, we found that NEAT1 could sponge miR-133a, and miR-133a could target the 3′-UTR of ARPC5. Therefore, we proposed and tested the hypothesis that NEAT1 might regulate MM progression through the miR-133a/ARPC5 axis. Through this study, we hope to provide a reference for further understanding of the NEAT1 regulated MM malignant process.
Material and methods
Sample collection
In this study, 20 MM patients and 20 healthy normal donors (iron deficiency anemia patients, due to menorrhagia or hemorrhoid blood loss, and excluding malignant tumors) were recruited at the First Affiliated Hospital, Hengyang Medical School, University of South China. Bone marrow samples were obtained from each participant. All subjects signed written informed consent. Our study was approved by the Ethics Committee of the First Affiliated Hospital, Hengyang Medical School, University of South China.
Cell culture and transfection
Four MM cell lines (NCl-H929, MM1S, U266, and RPMI-8226) and normal plasma cells (nPCs) were purchased from ATCC (Manassas, VA, USA), while OPM2 cells (an MM cell line) were obtained from Biovector (Beijing, China). All cell lines were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) plus 10% FBS (Gibco) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) at 37°C with 5% CO2. Lentivirus short hairpin RNA against NEAT1 or ARPC5 (sh-NEAT1 or sh-ARPC5), miR-133a mimic or inhibitor (miR-inhi), and their negative controls (sh-NC, mimic NC and inhi-NC) were synthesized by RiboBio (Guangzhou, China). They were transfected into NCl-H929 cells by Lipofectamine 3000 (Invitrogen).
Quantitative real-time PCR (qRT-PCR)
RNA was isolated by TRIzol reagent (Invitrogen), and cDNA was obtained using a PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). Then, cDNA was used for qRT-PCR by SYBR Green (TaKaRa) with specific primers (Table I). Fold change was analyzed by the 2-ΔΔCt method, and GAPDH or U6 served as an internal control.
Table I
Primer sequences used for qRT-PCR
Cell counting kit 8 (CCK-8) assay
Transfected NCl-H929 cells were seeded into 96-well plates and cultured overnight. Afterwards, cells were hatched with CCK-8 solution (Beyotime, Shanghai, China) at indicated time points for 4 h. The optical density (OD) values were determined at 450 nm using a microplate reader.
Soft-agar colony formation assay
Transfected NCl-H929 cells were plated into 6-well plates containing complete medium with 0.7% and 0.35% melted soft agar in the lower and upper layers, respectively. After 2 weeks, the stained colonies were imaged and counted using a microscope with AlphaView software.
Flow cytometry
Basing on the instructions of the Annexin V-FITC Apoptosis Detection kit (Beyotime), transfected NCl-H929 cells were harvested and suspended with binding buffer followed by staining with Annexin V-FITC and propidium iodide. Finally, the cell apoptosis rate was detected using a flow cytometer.
Dual-luciferase reporter assay
The partial sequences of NEAT1 or ARPC5 3′-UTR containing the binding sites or mutant sites of miR-133a were inserted into the pmirGLO vector to generate NEAT1-wt/mut or ARPC5-wt/mut vectors. Generated vectors and the miR-133a mimic or mimic-NC were co-transfected into NCl-H929 cells. Relative luciferase activity was detected by the Dual-Lucy Assay Kit (Solarbio, Beijing, China) after 48 h.
Fluorescence in situ hybridization (FISH) assay
NCl-H929 cells were seeded on an 8-well chamber slide overnight and fixed with 4% paraformaldehyde. After incubated with Triton X-100, cells were incubated with a biotin-labeled NEAT1 probe and FAM-labeled miR-133a probe (RiboBio) overnight. Cell nuclei were stained with DAPI, and then the fluorescence image was photographed by confocal microscopy.
Western blot analysis
Total protein was extracted by RIPA buffer, separated through 10% SDS-PAGE gel, and transferred onto PVDF membranes. After blockage, the membrane was incubated with anti-ARPC5 (ab51243, 1 : 5000, Abcam, Cambridge, MA, USA) or anti-GAPDH (ab9485, 1 : 2500, Abcam) followed by incubation with secondary antibody (ab205718, 1 : 50000, Abcam). Protein signals were examined by ECL Western Blotting Substrate (Solarbio), and the gray value was analyzed by Image-Pro Plus software.
Animal experiments
The BALB/c nude mice (thymus free mice, widely used in the study of immunology, oncology and disease pathogenesis) (Vital River, Beijing, China) were randomly divided into two groups: the sh-NC group and the sh-NEAT1 group (6 mice in each group). NCl-H929 cells transduced with sh-NEAT1/sh-NC were implanted into BALB/c nude mice through subcutaneous injection. Tumor volume was measured weekly. After 35 days, mice were euthanized, and tumor tissues were weighed. For immunohistochemical (IHC) staining, tumor tissues were cut into 4-µm thickness slides. After dewaxing, rehydration, and antigen retrieval, the slides were incubated with Ki-67 antibody (ab15580, 1 : 200, Abcam) and secondary antibody (ab150081, 1 : 1000, Abcam). The sections were stained with DAB solution and hematoxylin solution, and then the Ki-67 positive cells were observed by light microscopy. Animal studies were approved by the Animal Ethics Committee of the First Affiliated Hospital, Hengyang Medical School, University of South China.
Statistical analysis
GraphPad 7.0 software was applied for data analysis. Data are shown as means ± SD. The independent samples t-test was used for comparison between 2 groups. Analysis of variance (ANOVA) was used for comparison between multiple groups, and Tukey’s multiple comparisons test was used for post hoc comparison. Correlation analysis was performed by Pearson correlation analysis. Statistical significance was set at p < 0.05.
Results
NEAT1 knockdown suppressed MM cell growth
In the bone marrow specimens of MM patients, NEAT1 expression was found to be higher than that in healthy normal donors (Figure 1 A). High NEAT1 expression was also detected in five MM cell lines (NCl-H929, MM1S, U266, RPMI-8226 and OPM2) compared to nPCs (Figure 1 B). To further explore the role of NEAT1 in MM progression, sh-NEAT1 was transfected into NCl-H929 cells to silence NEAT1 (Figure 1 C). The results of CCK-8 assay indicated that NEAT1 knockdown significantly inhibited NCl-H929 cell viability (Figure 1 D). The colony numbers of NCl-H929 cells also could be reduced by transfection of sh-NEAT1 (Figure 1 E). The results of apoptosis rate detection revealed that NEAT1 silencing markedly enhanced the apoptosis of NCl-H929 cells (Figure 1 F). These data suggested that NEAT1 might accelerate MM cells proliferation and inhibit apoptosis.
Figure 1
Effects of NEAT1 knockdown on MM cell growth. A – NEAT1 expression in the bone marrow specimens from MM patients and healthy normal donors was measured by qRT-PCR. B – qRT-PCR was used to examine NEAT1 expression in MM cells and nPCs cells. C–F – NCl-H929 cells were transfected with sh-NC or sh-NEAT1. C – NEAT1 expression was detected by qRT-PCR. D – CCK-8 assay was used to assess cell viability. E – Soft-agar colony formation assay was performed to evaluate colony numbers. **p < 0.01, ***p < 0.001. F – Flow cytometry was utilized to determine the cell apoptosis rate. The cell experiment was repeated 3 times. **p < 0.01, ***p < 0.001.
NEAT1 served as miR-133a sponge in MM
To complete the mechanism of NEAT1 regulated MM progression, we predicted the downstream miRNA of NEAT1 by StarBase software. MiR-133a was screened for binding sites with NEAT1, and we constructed the NEAT1-wt and NEAT1-mut vectors according to their binding sequences (Figure 2 A). Dual-luciferase reporter assay results indicated that miR-133a only restrained the luciferase activity of NEAT1-wt vector rather than the NEAT1-mut vector (Figure 2 B). FISH results revealed that both NEAT1 and miR-133a were co-localized in the cytoplasm of NCl-H929 cells (Figure 2 C). MiR-133a expression could be increased by sh-NEAT1, suggesting that miR-133a was negatively regulated by NEAT1 (Figure 2 D). Also, miR-133a was downregulated in the bone marrow specimens of MM patients, and its expression was negatively correlated with NEAT1 expression (Figures 2 E, F). The above data confirmed that NEAT1 sponged miR-133a in MM.
Figure 2
NEAT1 served as miR-133a sponge in MM. A – The sequences of NEAT1-wt and NEAT1-mut are shown. B – Dual-luciferase reporter assay was used to confirm the interaction between NEAT1 and miR-133a. C – FISH assay demonstrated the co-localization of NEAT1 and miR-133a in NCl-H929 cells. D – qRT-PCR was performed to examine miR-133a expression in NCl-H929 cells transfected with sh-NC or sh-NEAT1. E – MiR-133a expression was measured by qRT-PCR in the bone marrow specimens from MM patients and healthy normal donors. F – Pearson correlation analysis was used to examine the correlation between NEAT1 and miR-133a expression in the bone marrow specimens from MM patients. The cell experiment was repeated 3 times. **p < 0.01
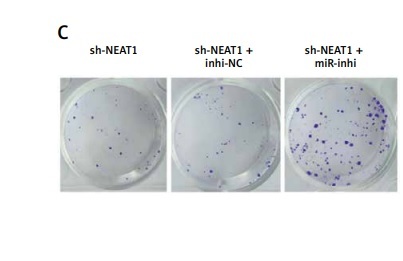
MiR-133a inhibitor reversed the effect of sh-NEAT1 on MM cell growth
To explore whether NEAT1 regulated MM progression via sponging miR-133a, rescue experiments were performed. After sh-NEAT1 and miR-inhi were co-transfected into NCl-H929 cells, the sh-NEAT1-mediated increase of miR-133a expression was abolished by miR-inhi (Figure 3 A). The addition of sh-NEAT1 + miR-inhi significantly enhanced the viability and colony numbers of NCl-H929 cells compared to the sh-NEAT1 + inhi-NC group (Figures 3 B, C). The promotion effect of NEAT1 knockdown on the apoptosis of NCl-H929 cells also was eliminated by the miR-133a inhibitor (Figure 3 D). These results indicated that NEAT1 indeed targeted miR-133a to promote MM progression.
Figure 3
Effects of sh-NEAT1 and miR-133a-inhi on MM cell growth. NCl-H929 cells were transfected with sh-NEAT1, sh-NEAT1 + inhi-NC or sh-NEAT1 + miR-inhi. A – MiR-133a expression was detected by qRT-PCR. B – Cell viability was examined by CCK-8 assay. C – Colony numbers were determined by soft-agar colony formation assay. D – Cell apoptosis rate was analyzed by flow cytometry. The cell experiment was repeated 3 times. **p < 0.01
ARPC5 was targeted by miR-133a in MM
StarBase software also was used to predict the target of miR-133a, and the 3′-UTR of ARPC5 was found to bind complementally to miR-133a (Figure 4 A). Dual-luciferase reporter assay results further confirmed the interaction between them, showing that only the luciferase activity of the ARPC5-wt vector was reduced by the miR-133a mimic (Figure 4 B). ARPC5 expression was promoted by miR-133a inhibition and inhibited by miR-133a overexpression at the mRNA level and protein level (Figures 4 C, D). In the bone marrow specimens of MM patients, ARPC5 was markedly upregulated and was negatively correlated with miR-133a expression, while it was positively correlated with NEAT1 expression (Figures 4 E–G). All data verified that miR-133a targeted ARPC5 in MM.
Figure 4
ARPC5 was targeted by miR-133a in MM. A – The sequences of ARPC5-wt and ARPC5-mut are presented. B – Dual-luciferase reporter assay was performed to confirm the interaction between miR-133a and ARPC5. C, D – qRT-PCR and western blot analysis were used to examine ARPC5 mRNA and protein expression in NCl-H929 cells transfected with miR-inhi or miR-133a mimic. E – ARPC5 expression was detected by qRT-PCR in the bone marrow specimens from MM patients and healthy normal donors. **p < 0.01. F, G – Pearson correlation analysis was employed to analyze the correlation between ARPC5 and miR-133a or NEAT1 expression in the bone marrow specimens from MM patients. The cell experiment was repeated 3 times. **p < 0.01
NEAT1 regulated the miR-133a/ARPC5 axis to mediate MM cell growth
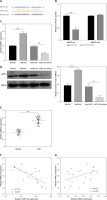
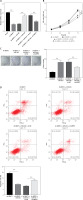
Further analysis was performed to confirm whether NEAT1 regulated MM progression through the miR-133a/ARPC5 axis. NCl-H929 cells were co-transfected with sh-NEAT1, miR-inhi and sh-ARPC5. MiR-133a inhibitor reversed the decreasing effect of NEAT1 knockdown on ARPC5 expression, and this effect was abolished by the addition of sh-ARPC5 (Figure 5 A). Downregulated ARPC5 eliminated the promotion effect of the sh-NEAT1 + miR-inhi + sh-NC group on the viability and colony numbers of NCl-H929 cells (Figures 5 B, C). Moreover, the inhibitory effect of the sh-NEAT1 + miR-inhi + sh-NC group on the apoptosis of NCl-H929 cells was also overturned by ARPC5 knockdown (Figure 5 D). These results showed that NEAT1 sponged miR-133a to positively regulate ARPC5, thereby promoting MM cell proliferation and suppressing apoptosis.
Figure 5
NEAT1 affects MM cell growth by regulating miR-133a/ARPC5 axis. NCl-H929 cells were transfected with sh-NC, sh-NEAT1, sh-NEAT1 + inhi-NC, sh-NEAT1 + miR-inhi, sh-NEAT1 + miR-inhi + sh-NC or sh-NEAT1 + miR-inhi + sh-ARPC5. A – ARPC5 expression was detected by qRT-PCR. B – CCK-8 assay was employed to assess cell viability. C – Colony numbers were detected using soft-agar colony formation assay. **p < 0.01. D – Flow cytometry was used to determine the cell apoptosis rate. The cell experiment was repeated 3 times. **p < 0.01.
NEAT1 silencing could inhibit MM tumor growth
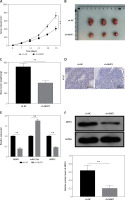
Animal experiments were performed to investigate the role of NEAT1 in MM tumor growth in vivo. After 35 days, we found that the tumor volume, size and weight were markedly reduced in the sh-NEAT1 group (Figures 6 A–C). Moreover, proliferation marker Ki-67 positive cells were significantly decreased in the tumor tissues of the sh-NEAT1 group (Figure 6 D). Additionally, we confirmed that NEAT1 and ARPC5 levels were decreased, while miR-133a level was increased in the tumor tissues of the sh-NEAT1 group (Figures 6 E, F). All the data indicated that NEAT1 might accelerate MM tumor growth by the miR-133a/ARPC5 axis in vivo.
Figure 6
Effects of NEAT1 on MM tumor growth. NCl-H929 cells transfected with sh-NC or sh-NEAT1 were injected into mice. Tumor volume (A), size (B) and weight (C) were measured in each group. D – Ki-67 positive cells in the tumor tissues of each group were analyzed by IHC staining. E, F – NEAT1, miR-133a and ARPC5 expression levels in the tumor tissues of each group were determined by qRTPCR. F – Western blot analysis was used to detect ARPC5 protein expression in the tumor tissues of each group. N = 6, **p < 0.01
Discussion
The development of human diseases is a complex process, which is accompanied by the abnormal expression of many genes. Here, we selected NEAT1 to explore its role and molecular mechanism in the MM process. Many studies have shown that NEAT1 acts as a proto-oncogene to promote tumorigenesis, including colorectal cancer [22] and papillary thyroid carcinoma [23]. NEAT1 has been confirmed to be upregulated in MM patients and its overexpression might contribute to MM cell proliferation and the cell cycle through the activation of PI3K/AKT signaling [24]. Previous studies have reported that the overall survival of MM patients was significantly reduced in patients with high expression of NEAT1 [25, 26]. Furthermore, resveratrol played an anti-tumor role in MM progression by reducing NEAT1 expression [27]. Therefore, it is reasonable to believe that NEAT1 also plays a carcinogenic role in MM progression. In our research, we confirmed the high expression of NEAT1 in MM patients and cell lines. Silencing of NEAT1 restrained MM cell proliferation and accelerated apoptosis, as well as inhibiting MM tumor growth in vivo, which was consistent with previous research [24, 27]. These results revealed that targeting NEAT1 might be an effective strategy for MM treatment.
Using bioinformatics analysis, we indicated that NEAT1 might interact with miR-133a. MiR-133a has been proved to be underexpressed in many tumor tissues and to negatively regulate tumorigenesis [28]. A previous study showed that miR-133a suppressed cell proliferation and metastasis to inhibit colorectal cancer progression [29]. Similarly, miR-133a was also found to play an anti-tumor role in lung cancer, which could hinder cell growth and metastasis [30]. Further analysis confirmed that NEAT1 and miR-133a were mainly localized in the cytoplasm, and NEAT1 could serve as a miR-133a sponge. Our data showed that miR-133a also had low expression in MM tissues and its expression was negatively correlated with NETA1 expression. The MiR-133a inhibitor overturned the inhibitory effect of sh-NEAT1 on MM cell growth, which not only confirmed that NEAT1 sponged miR-133a to facilitate MM progression, but also revealed the tumor suppressive role of miR-133a in MM, which was consistent with its role in other tumors [28–30].
Further studies revealed the complementary binding sites between ARPC5 and miR-133a. ARPC5 was considered to be upregulated in hepatocellular carcinoma and could be used as an independent biomarker to predict poor prognosis in patients [31]. miR-141 reduced tumor regeneration and metastasis in prostate cancer by targeting ARPC5 [32]. Knockdown of ARPC5 has also been shown to inhibit head and neck squamous cell carcinoma cell metastasis [33]. Previous studies reported that high expression of ARPC5 in MM was associated with poor overall survival of MM patients [21]. Consistent with this study [21], we further verified that ARPC5 showed elevated expression in MM patients. Additionally, we confirmed that NEAT1 positively regulated ARPC5 expression, and ARPC5 knockdown counteracted the effect of miR-inhi on sh-NEAT1-mediated MM cell growth, which fully confirmed the conclusion that NEAT1 promoted MM progression through the miR-133a/ARPC5 axis.
However, there are some limitations of this study. Given the ongoing collection of patient-relevant prognostic information, the prognostic value of the NEAT1/miR-133a/ARPC5 axis in this study needs to be further analyzed in the future. In addition, the downstream pathway by which ARPC5 regulates MM progression remains unclear. We only analyzed the expression trend and correlation of the NEAT1/miR-133a/ARPC5 axis in patients with MM by collecting clinical samples, and did not directly verify the effect of the NEAT1/miR-133a/ARPC5 axis on the progression of MM at the clinical level. Therefore, the presence of the NEAT1/miR-133a/ARPC5 axis needs to be validated at the clinical level in the future.
In conclusion, our research showed that NEAT1 promoted MM cell growth depending on the regulation of the miR-133a/ARPC5 axis. These findings provide new evidence for NEAT1 as a potential therapeutic target for MM and help us to better understand the tumor-promoting effects of NEAT1 in MM.