Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
OBSTETRICS AND GYNAECOLOGY / CLINICAL RESEARCH
Profiles of circular RNAs in human blood and their potential roles in preeclampsia
1
Department of Obstetrics and Gynecology, Center for Translational Medicine, Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, West China Second University Hospital, Sichuan University, Chengdu, China
2
Center for Translational Medicine, Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, Department of Obstetrics and Gynecology, West China Second University Hospital, Sichuan University, Chengdu, China
Submission date: 2021-09-05
Final revision date: 2021-12-21
Acceptance date: 2022-01-14
Online publication date: 2022-02-18
KEYWORDS
TOPICS
ABSTRACT
Introduction:
To identify circular RNA (circRNA) expression profiles in the blood of preeclampsia (PE) and healthy pregnant women, and further clarify the possible mechanisms of circRNAs involved in the pathogenesis of PE.
Material and methods:
Whole blood samples were collected from 5 paired PE and healthy pregnant women. The differentially expressed circRNAs (DE-circRNAs) were investigated using high-throughput sequencing. Bioinformatics was performed to evaluate the sequencing results and obtain insights into possible mechanisms, such as gene ontology (GO) and KEGG pathway analyses. Then, six DE-circRNAs were chosen and validated by quantitative real-time PCR (RT-qPCR) in an enlarged sample size. Their diagnostic values were analyzed by the receiver operating characteristic (ROC) curve. Establishment and analysis of the circRNA-miRNA-mRNA network were performed for the validated DE-circRNAs.
Results:
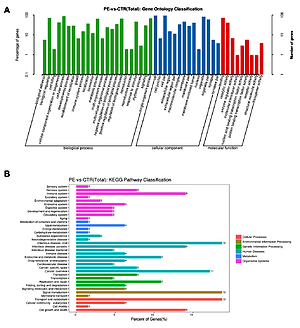
A total of 139 DE-circRNAs between PE and controls were revealed. Of them, 18 circRNAs were upregulated, and 31 circRNAs were downregulated (fold change > 2 and p-value < 0.05). Three circRNAs (has-circ-0007717, has-circ-0006460, and has-circ-0093055) were higher in both blood of early-onset and late-onset PE patients. The ROC analysis showed that the area under the curve values of has-circ-0007717, has-circ-0006460, and has-circ-0093055 were 0.64 (p = 0.11), 0.72 (p = 0.01), and 0.72 (p = 0.01), respectively. The circRNA-miRNA-mRNA competing for the endogenous RNA (ceRNA) network comprised 2 circRNAs, 154 miRNAs, and 6 mRNAs. KEGG analysis of these mRNAs included immune response-related pathways and cellular senescence.
Conclusions:
The CeRNA regulatory network indicated that DE-circRNAs might participate in the processes of the cell immune response.
To identify circular RNA (circRNA) expression profiles in the blood of preeclampsia (PE) and healthy pregnant women, and further clarify the possible mechanisms of circRNAs involved in the pathogenesis of PE.
Material and methods:
Whole blood samples were collected from 5 paired PE and healthy pregnant women. The differentially expressed circRNAs (DE-circRNAs) were investigated using high-throughput sequencing. Bioinformatics was performed to evaluate the sequencing results and obtain insights into possible mechanisms, such as gene ontology (GO) and KEGG pathway analyses. Then, six DE-circRNAs were chosen and validated by quantitative real-time PCR (RT-qPCR) in an enlarged sample size. Their diagnostic values were analyzed by the receiver operating characteristic (ROC) curve. Establishment and analysis of the circRNA-miRNA-mRNA network were performed for the validated DE-circRNAs.
Results:
A total of 139 DE-circRNAs between PE and controls were revealed. Of them, 18 circRNAs were upregulated, and 31 circRNAs were downregulated (fold change > 2 and p-value < 0.05). Three circRNAs (has-circ-0007717, has-circ-0006460, and has-circ-0093055) were higher in both blood of early-onset and late-onset PE patients. The ROC analysis showed that the area under the curve values of has-circ-0007717, has-circ-0006460, and has-circ-0093055 were 0.64 (p = 0.11), 0.72 (p = 0.01), and 0.72 (p = 0.01), respectively. The circRNA-miRNA-mRNA competing for the endogenous RNA (ceRNA) network comprised 2 circRNAs, 154 miRNAs, and 6 mRNAs. KEGG analysis of these mRNAs included immune response-related pathways and cellular senescence.
Conclusions:
The CeRNA regulatory network indicated that DE-circRNAs might participate in the processes of the cell immune response.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.