Biology research has developed considerably since next-generation sequencing (NGS) technology was created in 2005. Since then, NGS has been widely applied clinically while making a great contribution to the screening and diagnosis of cancer as well as rapid detection of infection [1]. Recently, chronic wounds associated with microbial infections have become a popular focus of clinical research. Traditionally, the detection of pathogens highly depends on laboratory techniques and conditions. Metagenomic next-generation sequencing (mNGS), as a novel method, is regarded as a promising approach to detect pathogens more rapidly and accurately. This technique can identify pathogens by analyzing DNA or RNA in target specimens. After removing the background gene data of human beings and according to the established microbial gene database, the results were obtained after big data analysis by integrating the type of specimens, the location of sampling, the patient’s medical history, blood picture changes, and other factors. The Illumina sequencing platform is often used, which is fast, accurate, and cost-effective [2].
At present, mNGS technology is quite effective in the diagnosis of diseases of the nervous system [3], diseases of the respiratory system [4], COVID-19 [5], diseases of the digestive system [6], and urinary system infection [7] through fluid samples. However, the sensitivity and specificity of mNGS through tissue samples, which has a powerful impact on clinical application, still need to be investigated. To explore the value of mNGS in etiological diagnosis and guidance on the treatment of chronic wound infections, we collected tissue samples during the operation of patients who suffered from nonhealing granulation wounds for more than 2 months from March 2019 to June 2020.
A study was performed from March 2019 to June 2020 and patients suffering from chronic ulceration with nonhealing granulation wounds for more than 2 months were enrolled in this research. The study was approved by the institutional board of Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences (NO. KY-Q-2021-056-01), and informed consent was obtained from all patients.
Approximately 1 cm3 granulation tissue in the deep wound was collected during the operation, followed by preservation of the sample tube with 3–5 ml of normal saline. Then, the sample was placed in an icebox and delivered to the biology company for pathogen detection by mNGS. Basic information such as age and sex, MRI characteristics of the wound, results of the T-cell spot test (T-SPOT), reads, the number of special sequences and duration of mNGS, duration of bacteria culture, and pathogens were recorded.
A 1 cm3 granulation tissue was collected in DNase tubes to identify the potential pathogens. DNA libraries were constructed through PCR amplification, and the qualified DNA libraries were sequenced on an Illumina platform. After the removal of low-quality and short (length < 40 bp) reads, Burrows-Wheeler Alignment was used for computational subtraction of human host sequences mapped to the human reference genome (hg38 and YH sequences), and high-quality sequencing data were generated. After the removal of low-complexity reads, the remaining data were classified according to four (bacteria, parasites, fungi, viruses) microbial genome databases. Finally, the multiparameters of species in microbial genome databases were calculated and exported. The results were interpreted by professionals with microbiology and clinical backgrounds [8].
Seven patients (3 males and 4 females) aged 10 to 57 years were enrolled in our study. The average duration of mNGS was 2 days. Mycobacteria were found through mNGS in 6 of 7 patients with 3 cases of Mycobacterium tuberculosis. One of the cases included a right groin injury. After non-rigorous acupuncture treatment, a sinus formed on the wound, which persisted and did not heal. After debridement and mNGS investigation, it was found to be a tuberculosis infection as well as rifampicin resistance. After 10 days of negative pressure treatment and effective anti-tuberculosis drugs, the wound was closed, and the medication was continued for 9 months. After a total of 12 months of follow-up, no recurrence was found (Figure 1).
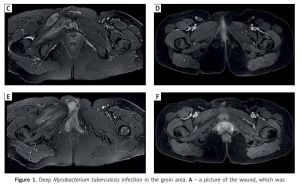
Figure 1
Deep Mycobacterium tuberculosis infection in the groin area. A – a picture of the wound, which was extensively granulomatous, upon admission. B – a picture of the wound 2 weeks after repair. C, E – representative magnetic resonance images (MRI) of the patient at the time of onset, showing lesions in the right groin and pelvic floor muscles. D, F – are MRI of the patient at 10 months after recovery, suggesting that the lesions in the groin and pelvic floor muscles had been absorbed
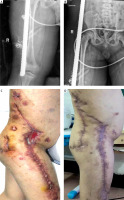
There were 2 cases of Mycobacterium abscessus, and 1 case of Mycobacterium fortuitum. One of the cases involved steel bars that were penetrated through the thigh and torso, and the wound did not heal after seven rounds of debridement. After 7 months of ineffective treatment, mNGS was used to first discover the Mycobacterium fortuitum infection. The wound was closed after using sensitive antibiotics for 9 days according to expert consensus. Medication continued for 10 months after reconstruction, and after 12 months of follow-up, no recurrence was found (Figure 2). Mycobacteria were also detected by bacterial culture in all 4 patients, which was consistent with the results of mNGS.
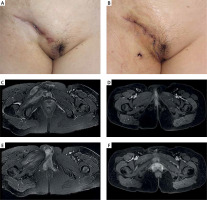
Figure 2
Incidental Mycobacterium infection after a thigh and trunk penetration injury. A, B – the X-ray anteroposterior views of the patient at the time of injury. C – the wound of the patient at 4 months after injury. D – the wound after pathogen analysis by mNGS and standard treatment for 5 months
In addition, coinfection of Staphylococcus aureus and cytomegalovirus was detected by mNGS in another patient. According to the results of mNGS, antibiotic and antivirus drugs were administered following expert consensus while the wound was debrided. All patients recovered successfully with no recurrence in a median follow-up period of 4 months. Patient information is summarized in Table I.
Table I
Characteristics of the patients
It has been demonstrated that most chronic wounds are closely associated with pathogen infections in the clinic. Rapid and accurate pathogen detection plays a key role in the treatment of patients with infections. Failed and delayed pathogen detection in the clinic can lead to prolonged hospitalization and rehospitalization with increased mortality up to 10% [9–11]. In addition, an empirical broad-spectrum antibiotic for patients with unknown pathogens will increase the risk of adverse effects and antibiotic resistance [12]. Currently, bacterial culture is still widely used for pathogenic detection in clinics. However, some special microorganisms, such as Treponema pallidum and Bartonella, could hardly be detected by bacterial culture, while mycobacteria and mold took several weeks to grow on the medium. As these special pathogens are difficult to detect quickly through conventional detection methods, novel detection approaches with properties of rapid detection and wider spectrum coverage are desirable for pathogen identification in the clinic [13]. mNGS technology has the natural advantages of rapid detection and relatively high sensitivity and specificity [8]. As a rapid diagnostic technique, mNGS has a wide range of applications regarding the microbiome, antimicrobial resistance [14], human host gene expression [15], and oncology. In addition to mNGS, accurate molecular testing through PCR is another available method for microbial identification. However, in comparison to mNGS, 16S rRNA PCR and sequencing are used for a limited number of pathogens at a time, or successful culture of microorganisms from clinical samples is required [8]. Moreover, 16S bacterial PCR and 28S-internal transcribed ribosomal gene spacer (28S-ITS) fungal PCR showed a higher false-negative rate and lower sensitivity in previous studies [16, 17].
The pathogens detected by mNGS can only represent the presence of these detected microorganisms in the specimen, but it cannot distinguish between colonizing, background, or pathogenic bacteria. Therefore, a modified version of the mNGS device was created. The provision of specimen collection sites could identify normal micro ecologies and the possible presence of opportunistic and colonizing bacteria. At the same time, combined with clinical data, such as current symptoms, changes in infection indicators (routine blood tests, C-reactive protein, procalcitonin, etc.), and imaging features are needed to make a comprehensive judgment. Moreover, a high number of detected microorganism sequences should be a priority for pathogenic pathogens [18].
However, due to the low nucleic acid extraction efficiency of intracellular infectious bacteria (tuberculosis bacillus, legionella, etc.) and pathogens with thick cell walls (such as fungi), pathogenic pathogens should be considered even if the number of detected sequences is not high [19]). Therefore, it is possible to detect coinfection using a modified version of this mNGS device.
Generally, a fluid sample is required for mNGS to identify pathogens. The application of wound tissue samples has seldom been reported. In our research, we found that mNGS can also identify pathogens accurately through wound tissue specimens, which was useful for prescribing antibiotics in the clinic. Based on the results above, we suggest that mNGS is a potential tool for pathogen detection in chronic wounds with granulation tissue, especially for mycobacterial identification. Compared with conventional microbiological identification, mNGS takes advantage of faster detection with a duration of 2 days on average and more accurate and specific results of mycobacteria, which makes a great contribution to reasonable antibiotic administration and differential diagnosis between tuberculosis mycobacteria and non-tuberculosis mycobacteria.
We suggest that mNGS has the following advantages in comparison to conventional bacterial culture: 1) has a higher positive rate, especially for anaerobe, tuberculosis mycobacteria, and non-tuberculosis mycobacteria detection, 2) can be used for virus and rare pathogen identification, and 3) can identify multiple pathogens simultaneously through one sample. As a comprehensive direct detection method, mNGS may eventually replace culture, antigen detection and PCR methods in clinical microbiology [20]. Furthermore, mNGS will have a pivotal role in monitoring and tracking new disease outbreaks. As mNGS bases of rapid diagnostic platforms are distributed globally, it will be possible to detect and control infectious outbreaks at a much earlier stage, saving lives and lowering costs.
However, there are still some limitations to our research. First, the sample size is small, and more patients are required for further investigation. Second, in addition to granulation wounds, other kinds of wounds, including bone, acute, and muscle wounds, should also be investigated for pathogen detection through mNGS.
In conclusion, mNGS is a promising tool for microbial identification with properties of rapid detection and high specificity, providing a novel approach for the detection of mycobacteria in chronic granulation wound infections.