Introduction
Osteoporosis is an outcome of an imbalance between osteoclast bone resorption and osteoblast bone formation [1]. Osteoclasts originate from multipotent hematopoietic stem cells and are capable of bone resorption in the human body. These cells have the capacity to differentiate into monocytes and macrophages [2]. Several hormones along with cytokines are involved in the process of osteoclastogenesis via transcription factors which modulate the survival, functioning, proliferation and differentiation of osteoclasts [3–6]. Colony-stimulating factor-1 (CSF-1) and osteoclast differentiation factor (ODF) both are activated by T cells or are released by osteoblasts. Both CSF-1 and ODF are the main cytokines involved in osteoclastogenesis [7–9]. ODF can elevate the levels of nuclear factor of activated T-cell cytoplasmic (NFATC-1) [10]. Markers of osteoclasts such as cathepsin K and tartrate-resistant acid phosphatase (TRAP) have been recognized to have multiple sites by NFATC-1 [10–12]. Phosphoinositide 3-kinase (PI3K) is found to phosphorylate phosphoinositides. PI3K is responsible for generating D3-phosphoinositides through the involvement of proteins such as Akt and also promotes various cellular functions which include differentiation, mutagenesis, motility and survival. However, signaling pathways mediated by PI3K can be damped by PTEN and SHIP. SHIP is widely present in hematopoietic cells and is responsible for the hydrolysis of the 5′-phosphate group of PIP3 which is the main product of PI3K and hence regulates the expression of PI3K negatively [13]. The literature hence suggests involvement of PI3K in differentiation of osteoclasts [14]. It is also evidenced that the PI3K/AKT pathway is responsible for the regulation of osteoclast differentiation and survival [15, 16]. MicroRNAs (miRNAs) are non-coding RNAs measuring 20 to 22 nucleotides. MiRNAs are known to play a potential role in regulating various cellular processes at the post-transcriptional level [17]. miRs have been identified to act at every step of osteogenesis i.e., from bone development in the embryonic stage to the adult stage. miRs have been identified to regulate the functional activity, differentiation and growth of osteocytes, osteoblasts and osteoclasts [18]. Remodeling of bones is an uninterrupted process and requires the association of osteoblast and osteoclast activities [19]. Reports suggest involvement of various miRs in regulation of osteoblasts. MiRs also regulate the role of osteoblasts in proliferation, differentiation and bone formation [20, 21]. Looking into the potential role of various miRs in the process of bone formation and related processes, we investigated the role of some specific miRs in differentiation of osteoclasts and their role in osteoporosis.
Recently a study involving patients with osteoporosis found the expression of 754 different miRs among which miR-23b-3p and miR-140-3p were significantly upregulated [22]. miR-23b-3p belongs to cluster of miR-23 of the microRNA family, which are reported to be involved in osteoporosis and in regulation of differentiation of osteoblasts [23, 24]. miR-23b-3p has been found to be associated with several pathways in cellular physiology. MiR-23b-3p is a member of the intronic miR-23b/27b/24-1 cluster and is located at the q22.32 position on chromosome 9. The pre-miR-23b produces the mature miR-23b-3p and miR-23b-5p, which is the complementary strand [25]. Further, it is necessary to distinguish the miR-23b/27b/24-1 cluster from the miR-23a/27a/24-2 cluster. It has been reported that miR-23a, -27a and 24-2 act as oncogenes in several cancers. High expression of miR-23b-3p is observed in the differentiation of keratinocytes [26], skeletal muscle [27] and chondrocytes [28]; it also promotes angiogenesis [29] and blocks the progression cycle of endothelial cells [30]. miR-23b-3p is also reported to be involved in the process of bone formation [22]. In a recent study, miR-23b-3p was found to promote carcinogenicity by targeting the PTEN/PI3K/Akt pathway in pancreatic cancer [31]. However, the role of miR-23b-3p in differentiation of osteoclasts and the involved mechanism remain unexplored.
The aim of the present research was to study the involvement of miR-23b-3p in the process of differentiation of osteoclasts and also to find the involved pathway. In the present study we used CSF-1 and ODF induced osteoclasts and TIB-71 cells. As CSF-1 and ODF are both synthesized by osteoblasts and are significant cytokines for the process of osteoclastogenesis, we used osteoclasts which were induced by CSF-1 and ODF for the study. TIB-71 cells are a RAW and monocyte/macrophage cell line which is suitable for transfection of the host. The study evidenced that expression of miR-23b-3p was upregulated in the process of CSF-1 and ODF induced osteoclastogenesis. We also found that blockade of miR-23b-3p caused significant suppression of osteoclast activity. We confirmed that miR-23b-3p functions by targeting PTEN and activating the PI3k/Akt pathway. The results of an in vivo experiment in osteoclast specific miR-23b-3p transgenic mice showed suppression in PTEN activity, elevated osteoclast activity and decreased bone density.
Material and methods
Cell culture
CSF-1 and ODF induced osteoclasts were obtained from R&D Systems Inc., USA. The bone marrow cells were cultured in Modified Eagle’s medium (Thermo Fisher USA) supplemented with fetal bovine serum (FBS) (10%) (Thermo Fisher USA), penicillin (1%), streptomycin (1%) and CSF-1 (10 ng/ml) for 24 h. Bone marrow monocytes (BMMs) were subjected to culture in Modified Eagle’s medium supplemented with FBS (10%), CSF-1 (30 ng/ml) and ODF (50 ng/ml). The TIB-71 macrophage cells (ATCC USA) were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher USA) supplemented with FBS (10%), streptomycin and penicillin (1%). The TIB-71 cells were transfected with ODF (50 ng/ml) for osteoclastogenesis. The cells were incubated under controlled conditions with 95% humidity and 5% CO2. The medium was replaced with a fresh one after every 48 h. The TIB-71 cells were transfected with miR-23b-3p mimics, anti-miR-23b-3p and negative control (NC) using Lipofectamine 2000 reagent (Thermo Fisher USA) for 5 days (mimics and anti-miR) and 2 days (NC). For blocking the expression of PI3K, the TIB-71 cells were treated with PI3K inhibitor LY-294,002 hydrochloride (40 mM) (Sigma-Aldrich USA), also called an autophagy inhibitor, for 24 h.
Extraction of RNA and microarray analysis
The isolation of RNA of TIB-71 cells previously induced with or without ODF was done. The miRs were extracted and labeled as Cy-3 or Cy-5. Microarray analysis was done using the miR database from TargetScan (version 7.1) using the source http://www.targetscan.org/vert_72/. The intensity of color was assessed using the reading program. The mean values of spots with repeating colors were used for analysis. The spots were normalized using the invariant set normalization technique and the intensities were converted to gene expression log-2 ratios (ratio of treatment groups and the control).
TRAP staining
The ODF induced cells were subjected to TRAP staining using the Acid Phosphatase kit (Thermo Fisher USA) following the supplied instructions. Briefly, after incubating the ODF induced cells for 3 days, the cells were fixed by soaking them in fixing solution (Sigma-Aldrich USA) for 20 s. The cells were rinsed with distilled water and were then stained with TRAP stain followed by incubation at room temperature under darkness for 1 h. The TRAP solution was removed by washing with distilled water. The TRAP-positive cells were viewed and recorded using a microscope (ZEISS, Germany).
Bone resorption pit assay
For the bone resorption pit analysis, the bone marrow monocytes (5 × 105 cells/well) were incubated in slices of bovine bones in a 24-well plate using the NeuroCult-XF Proliferation Media (Stem Cell Technologies) for 24 h and then switched to differentiation media for the next 72 h. The bovine bone sections were immersed in 1 molar ammonium hydroxide and sonicated for removal of adhered cells, after which the sections were stained using Toluidine blue solution (0.1%). Pit area against the total bone area of each section was recorded using image processing software from Media cybernet. Inc.
Extraction of total miRNA
The miRNeasy kit (Qiagen USA) was used for isolating total RNA from the isolated cells as per the supplier’s instruction. Briefly, the cells were harvested and subjected to lysis using lysis buffer (700 μl) and were then mixed with chloroform (140 μl). The cells in the tube were centrifuged at 10000 rpm for 10 min at 4ºC, then the supernatant consisting the aqueous layer was transferred to a mini spin column and then mixed with ethanol (100%). The collected total RNA was subjected to real-time PCR analysis.
Quantitative real-time PCR (qRT-PCR) and reverse transcription
For reverse transcription the bone tissues were processed to extract total RNA using TRIzol reagent (Invitrogen, USA). For the same, RNA (500 μg) was subjected to reverse transcription with Prime-Script RT reagent Kit (TaKaRa) following the provided instructions. For quantification of miR-23b-3p, stem-loop RT-PCR was used; about 2 μl of cDNA was utilized for detecting expression of mRNA and miRNA by qRT-PCR using Tli RNaseH Plus (TaKaRa). GAPDH was used as a loading control for mRNA and U6 for miRNA. Primers used for the study were obtained from Beijing Sunbiotech Co. China. The sequences of primers are depicted in Table I.
Table I
Sequences of primers used in the study
Luciferase reporter assay
TIB-71 cells were transferred to 6-well plates (1 × 106 cells /well). The cells received transfection of pGL3-PTEN 3′-UTR and pRL-TK (Promega USA), Renilla luciferase plasmids with anti-miR-23b-3p or NC (Lipofectamine, USA) and miR-23b-3p (Invitrogen USA); the transfection was done according to supplied instructions. The luminescence was measured using a luminometer (Promega), and the obtained values were normalized by Renilla luciferase assay.
Immunoblotting analysis
For expression of proteins the cells were lysed using buffer (Thermo Fisher USA) for 30 min. The cells were centrifuged at 10000 rpm at 4ºC for 20 min and the protein fractions were collected and subjected for SDS-PAGE followed by transfer to polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were blocked with nonfat dry milk and incubated with specific antibodies for 12 h. Secondary antibody labeled with horseradish was added and was analyzed using a chemiluminescence kit (Thermo Fisher USA). The antibodies against Akt, P-Akt, PTEN, NFATC-1, STAT3 and GAPDH (Santa Cruz biotech USA) were used for studying protein levels. GAPDH was used as a loading control for measuring the relative intensities of protein bands.
Generation of osteoclast-specific miR-23b-3p transgenic mice
Before the study all the animal experiments received approval by the animal ethical committee of Linyi People’s Hospital, China. The institutional ethical board sanctioned the animal protocols; the approval number was SY/2050214. The mouse Acp5 plasmid was constructed in the laboratory of Linyi People’s Hospital, China. The plasmid was constructed in pGL3 vector (Promega) by transfecting the promoter region from pKB5 (gifted by Dr. Jiwei Chai); the transfer was done with the help of Qucik change kit (Stratagene). The Acp5 plasmid, which is a promoter for osteoclast targeted gene expression, was transfected into mice to generate the osteoclast-specific miR-23b-3p transgenic mice (OC-TG23b). The mice were sub-cloned with pre-miR-23b-3p cDNA into a vector and substantiated the levels of miR-23b-3p in TIB-71 cells by PCR. Acp5-pre-miR23b-3p plasmid was created by introducing Xba/Kpn and the fragment of pre-miR-23b-3p into the responsible sites downstream of the Acp-5 promoter. The Acp-5-pre-miR-23b-3p fragments were injected in the mouse oocytes (C57BL/6J) and then they were transferred to the pseudo-pregnant mice. The resultant transgenic mice were subjected to breeding for up to 5 generations for obtaining mice of defined genetic make.
Cell sorting by flow cytometry
Bone marrow cells from both mice strains, i.e. OC-TG23b and the wild type (WT), were collected from the femur and tibia bone and were sorted using a flow cytometer (Thermo Fisher, USA). After collection, the bone marrow cells were washed using 1% phosphate buffer saline (PBS), after which they were stained with mouse OSCAR antibody followed by staining with IgG-PE antibody. The stained cells were subjected to FACS analysis. The resultant cell population was used for extraction of total RNA followed by qRT-PCR analysis.
Micro CT analysis
Ex vivo scanning of the femur of every mouse was done using the Quantum GX microCT system (PerkinElmer USA). Briefly, the obtained sections of bone (10 mm in diameter) were scanned specifically at the region of the proximal tibia extending until the tibia diaphysis. About 70 continuous sections starting from the extreme proximity of the growth plate were selected for the study. Every bone section was selected for a three-dimensional view for calculating various parameters.
Ethics
Ethics approval and consent to participate All the animal protocols were in accordance with the draft of the Animal protection law of the People’s Republic of China-2009 for experimental animals, the study received prior approval from the institutional ethical committee of Linyi People’s Hospital, China, and the approval number was SY/2050214.
Statistical analysis
All the results are presented as mean ± % relative standard deviation for every experiment. Equality of variance across every group was assessed; if found unequal we assorted the data showing a heterogeneous pattern for two-way or one-way analysis. In the lack of such conditions we selected a linear model comprising Student’s t-test. The p-values less than 0.05 and 0.01 were regarded as significant and highly significant respectively. All the analysis was done using GraphPad Prism software.
Results
miR-23b-3p is highly over-expressed in osteoclastogenesis induced by treatment with CSF-1 and ODF
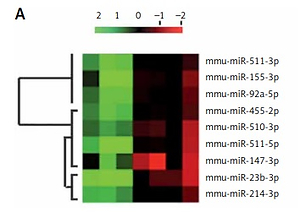
Microarray analysis (Qiagen, USA) was done in TIB-71 cells to analyze the involved miRNA in osteoclastogenesis. The osteoclast precursor cells were stimulated by treating with ODF; the cells without ODF were termed as controls. The findings of the experiment suggested that about 9 miRs were variably expressed during the process of differentiation of TIB-71 cells into osteoclasts (Figure 1 A). We evidenced that 4 miRs (miR-23b-3p, miR-214-3p, miR-510-3p and miR-155-3p) were significantly over-expressed, whereas 5 miRs (miR-147-3p, miR-511-5p, miR-455-2p, miR-92a-5p, miR-147-3p) were down-regulated. Further RT-PCR analysis was done to confirm the over-expression of 4 miRs; the findings were in agreement with those of miR microarray (Figure 1 B). Among them, miR-23b-3p was the most distinctively over-expressed, i.e., about 2 times compared to its control.
Figure 1
miR-23b-3p was over-expressed in osteoclastogenesis. A – Microarray analysis was done in TIB-71 cells with or without induction of ODF, the value of 2 green and red suggests low and high expression levels respectively. The microarray showed miRNAs which showed changes more than 1 to 5 fold. B – qRT-PCR analysis for the relative levels of miR in ODF-induced TIB-71 cells, the levels were normalized against U6 as loading control. C – The bone marrow monocytes (BMMs) were cultured in CSF-1 (30 ng/ml) and ODF (50 ng/ml) for the mentioned time period. The qRT-PCR analysis was done for estimating relative levels of miR-23b-3p in CSF-ODF-treated BMMs. The levels of miR-23b-3p were normalized against U6 as a loading control. D – The qRT-PCR analysis for relative mRNA levels of ACP-5 miR in CSF-ODF-treated BMMs and were normalized against GAPDH. E – Expression levels of TRAP protein in BMMs after treating them with ODF were studied by western blot analysis; GAPDH was selected as a loading control
All the results are presented as mean ± SEM (n = 3). **P < 0.01 compared to control, #p < 0.01 compared to day 0, @p < 0.01 compared to day 3 after treatment.
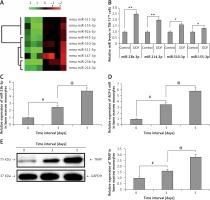
During the process of ODF and CSF-1 induced osteoclastogenesis from bone marrow monocytes the levels of miR-23b-3p increased progressively with time (p < 0.01) (Figure 1 C). Briefly, the expression of miR-23b-3p started to elevate progressively on the 3rd day post induction, and on the 5th day the levels were at least 6 times higher compared to that on the 1st day. qRT-PCR analysis for the levels of Acp5, i.e. osteoclast effector gene, and TRAP in CSF-1 and ODF induced bone marrow monocytes suggested that the changes in expression levels of miR-23b-3p were in agreement with the expression of Acp5 and TRAP (Figures 1 D, E). The outcomes suggested that miR-23b-3p plays a potential role in osteoclastogenesis. The expression of genes was normalized against GAPDH as a loading control.
miR-23b-3p is crucial to osteoclastogenesis
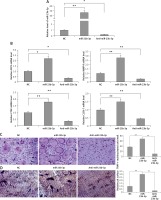
To evaluate the significance of miR-23b-3p for differentiation of osteoclasts, the bone marrow monocytes were treated with anti- or miR-23b-3p mimics in the process of osteoclastogenesis. The outcomes showed that the cellular levels of miR-23b-3p were significantly overexpressed by treatment of miR-23b-3p mimics and downregulated when treated with anti-miR-23b-3p (Figure 2 A). The levels of MMP-9, TRAP, cathepsin K and CLC-7 are potential markers suggesting the process of differentiation and activity of osteoclast [32, 33]. The results showed that, after exposure of 5 days, the levels of ACP-5, CTS-K, MMP-9 and CLC-7 were significantly upregulated after treating with miR-23b-3p mimics and downregulated when treated with anti-miR23b-3p against negative control (Figure 2 B). We observed that the TRAP positive osteoclasts with multiple nuclei per well were significantly increased in cells treated with miR-23b-3p mimics and were reduced when they received treatment of anti-miR-23b-3p against the negative control group (Figure 2 C). Also, due to the suppressed osteoclast activity the relative surface of osteoclast bone-resorbing pits also decreased in the group receiving treatment with anti-miR-23b-3p (Figure 2 D). The outcomes showed that miR-23b-3p is vital for osteoclastogenesis.
Figure 2
miR-23b-3p regulates osteoclastogenesis in bone marrow monocytes. A – BMMs treated with CSF-1 and ODF were subjected to qRT-PCR for analysis of miR-23b-3p after treating them with miR-23b-3p mimic or anti-miR-23b-3p or negative control (NC); the levels were normalized against U6 as loading control. B – qRT-PCR analysis was done to evaluate relative mRNA levels of MMP9, ACP5, CTSK and CLCN7; the levels were normalized using GAPDH as a loading control. C – Microscopic examination showing images of osteoclast formation; the images show that the TRAP-positive osteoclasts with multiple nuclei in cells treated with miR-23b-3p mimics, anti-miR-23b-3p and negative control group. D – Image analysis study was done for studying the areas showing bone resorption on the bone slices
*P < 0.05, **P < 0.01 compared to negative control.
PTEN is the direct target of miR-23b-3p
In silico analysis suggested that the 3′-UTR region of PTEN had a miR-23b-3p binding site. Previously, miR-23b-3p has been identified to have a binding site on the 3′-UTR region in pancreatic cancer cells [34]. We used two algorithms, i.e. CHIP base and the miRecords; the data indicated the predicted number of target genes regulated by miR-23b-3p. One of the target genes, PTEN, was reported previously to a play major role in ODF-induced differentiation of osteoclasts [35]; hence among the list of genes regulated by miR-23b-3p it was chosen for further evaluation. The results suggested that the 3′-UTR region of mouse PTEN had a putative region (nucleotides 3102-3109) which was common with the sequence of mature miR-23b-3p (Figure 3 A). To evaluate whether PTEN was a favorable target of miR-23b-3p, we studied the effect of mimic-, anti- and NC miR-23b-3p on luciferase activity in the TIB-71 cells which received transfection of the 3′-UTR fragment of PTEN reporter. The outcomes showed that miR-23b-3p significantly inhibited the luciferase activity of the 3′-UTR fragment of PTEN; we also found that the luciferase activity increased significantly after suppressing the endogenous miR-23b-3p levels after treating with anti-miR-23b-3p (Figure 3 B). We also evaluated the effect of miR-23b-3p and anti-miR-23b-3p on the mRNA and protein levels of PTEN in the ODF-induced TIB-71 cells. The results suggested that miR-23b-3p mimic caused a dip in levels of PTEN protein in ODF induced TIB-71 cells, whereas anti-miR-23b-3p caused an increase in protein levels of PTEN compared to the negative control (Figure 3 C), and we also noticed that there were no significant changes in PTEN mRNA levels (Figure 3 D).
Figure 3
PTEN in osteoclasts is the direct target of miR-23-3p. A – Sequence showing binding sites between mouse miR-23b-p with 3′-UTR region of PTEN. B – Effect of miR-23b-3p, anti-miR-23b-3p and negative control on luciferase activity in TIB-71 cells transfected with 3′-UTR reporter of PTEN. C – Effect of miR-23b-3p, anti-miR-23b-3p and negative control on protein levels of PTEN. miR-23b-3p mimic, anti-miR-23b-3p or negative control were transfected in ODF-induced TIB-71 cells. For the study GAPDH was selected as a loading control. D, E – qRT-PCR analysis for mRNA levels of PTEN. For the study GAPDH was selected as a loading control. The expression of STAT3 was assessed by immunoblotting analysis; GAPDH was selected as loading control
*P < 0.05, **p < 0.01 compared to negative control, NS – not significant.
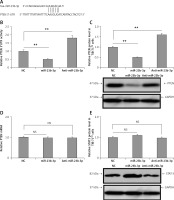
One more gene STAT3 was among the listed genes regulated by miR-23b-3p; hence after PTEN we studied any possible involvement of STAT3 in the process of differentiation of osteoclasts. However, we found no changes in expression levels of STAT3 after the transfection of mimics in the TIB-71 cells induced by ODF (Figure 3 E). These outcomes clearly suggest that PTEN was the favorable target of miR-23b-3p in the process of osteoclast differentiation.
miR-23b-3p promotes differentiation of osteoclast via the PTEN cascade
Previously it has been reported that PTEN is involved in regulation of ODF-mediated osteoclast differentiation in TIB-71 cells, i.e. the precursor cells via the PI3K/Akt pathway [35]. To evaluate the effect of miR-23b-3p on the PTEN/AKT axis during the ODF-mediated osteoclast differentiation, the TIB-71 cells received treatment with ODF, ODF + anti-miR-23b-3p, ODF + miR-23b-3p mimics, and LY294002 (PI3K inhibitor) + miR-23b-3p mimics. The intracellular levels of miR-23b-3p were upregulated by ODF treatment and reduced by treatment with anti-miR-23b-3p. The PI3K inhibitor showed no changes in the intracellular levels of miR-23b-3p (Figure 4 A). We also found that the PI3K inhibitor blocked the expression of p-AKT and NFATC-1 in the TIB-71 cells (Figure 4 B); hence we analyzed the levels of PTEN, p-AKT, AKT and NFATC-1 in the TIB-71 cells (Figure 4 A). The outcomes showed that the protein levels of PTEN were down-regulated, and NFATC-1 and AKT were up-regulated after induction with ODF. In addition, anti-miR-23b-3p inhibited the levels of p-AKT and NFATC-1 via upregulating the expression of PTEN. The treatment of miR-23b-3p mimics suppressed the protein levels of PTEN in TIB-71 cells, whereas the levels of p-AKT and NFATC-1 were upregulated. The PI3K inhibitor potentially suppressed the miR-23b-3p mediated AKT activation (Figure 4 C). We also found that anti-miR-23b-3p and PI3K inhibitor blocked the elevated mRNA levels of ACP-5, CTSK, NFATC-1, CLCN-7 and MMP-9 mediated by ODF (Figure 4 D). All the data indicated that miR-23b-3p regulated the ODF-mediated activation of the PI3K/AKT signaling cascade responsible for differentiation of osteoclasts by targeting PTEN.
Figure 4
miR-23b-3p encourages differentiation of osteoclasts via PTEN pathway. A – qRT-PCR analysis was done for expression levels of miR-23b-3p in TIB-71 cells, after inducing them with ODF, miR-23b-3p, anti-miR-23b-3p and LY294002, the expression levels were normalized against U6. B – Effect of ODF and LY294002 on the protein levels of NFATC-1 and pAKT in TIB-71 cells; GAPDH was selected as a loading control. C – Effect of miR-23b-3p, anti-miR-23b-3p and LY294002 on the protein levels of NFATC-1, pAKT and AKT in TIB-71 cells; GAPDH was selected as a loading control. D – Effect of miR-23b-3p, anti-miR-23b-3p and LY294002 on mRNA levels of NFATC-1, ACP-5, MMP-9, CLCN7 and CTSK in TIB-71 cells; GAPDH was selected as a loading control
*P < 0.05, **p < 0.01, ##p < 0.01, @@p < 0.01,^^p < 0.01, NS – not significant.

miR-23b-3p promotes osteoclast activity in vivo
To evaluate the involvement of miR-23b-3p in vivo, we constructed a plasmid that expresses miR-23b-3p under the ACP-5 promoter. ACP-5 is reported to be over-expressed in osteoclasts [36]. At the same time, we developed ACP-5-miR-23b-3p transgenic mice (OC-TG23b-3p). We analyzed the expression of miR-23b-3p in OC-TG23b-3p and in the wild-type (WT) mice; we found that the OC-TG23b-3p mice presented significantly higher levels of miR-23b-3p in the bone tissues at the age of 8 weeks (Figure 5 A). The bone mineral density (BMD) was evaluated by microcomputed tomography (MCT). It was observed that the ratio of bone volume (BV) and the tissue volume (TV), i.e. BV/TV, in OC-TG23b-3p mice was significantly lower compared to the WT mice. The trabecular space (TS) in the OC-TG23b-3p mice was found to be significantly elevated compared to the WT (Figures 5 B, C). We also found that the mRNA levels of CTSK, NFATC-1, MMP-9, ACP-5 and CLCN-7 were significantly elevated in the osteoclast cells from the OC-TG23b-3p mice (Figure 5 D). TRAP staining for number of TRAP-positive multinucleated cells in the ODF-induced bone marrow monocytes isolated from the bone marrow of OC-TG23b-3p and wild type mice suggested that the formation of TRAP-positive osteoclasts was promoted in ODF-induced bone marrow monocytes from the OC-TG23b-3p mice (Figure 5 E). We also found that the osteoclast bone-resorbing activity increased due to upregulation of osteoclast formation (Figure 5 F). In addition, we isolated bone marrow monocytes from the bone marrow of both OC-TG23b-3p and WT mice and observed that the levels of PTEN after ODF induction were significantly lower in the bone marrow monocytes of OC-TG23b-3p mice compared to WT mice and decreased rapidly with ODF treatment. In association with this, the levels of p-AKT and NFATC-1 were increased in bone marrow monocytes from the OC-TG23b-3p compared to WT mice. After treating with ODF, the over-expression of the proteins was significantly higher in ODF-induced bone marrow monocytes OC-TG23b-3p compared to WT (Figures 5 G–J). The findings hence suggested that over-expression of miR-23b-3p in osteoclasts causes activation of osteoclast activity and bone loss in the animal model.
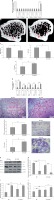
Figure 5
miR-23b-3p promotes osteoclast activity in vivo. A – qRT-PCR study was done to study the levels of miR-23b-3p in bone and other tissues; the levels were normalized against the levels of wild type mice and OC-TG23b-3p mouse lines; the levels were normalized against U6. B – The microCT analysis presented images in OC-TG23b and wild type mice. C – Micro CT-scan for evaluation of Tb.Sp, BMD and BV/TV in the tibular bone of wild type OC-TG23b-3p mice. D – qRT-PCR study was done for relative mRNA levels of NFATC-1, ACP-5, MMP-9, CTSK and CLCN-7 in osteoclasts separated by flow cytometry; the levels were normalized against GAPDH as a loading control. E – Results of TRAP staining for number of TRAP-positive multinucleated cells in the ODF-induced bone marrow monocytes isolated from the bone marrow of OC-TG23b-3p and wild type mice. F – Image analysis suggested areas of bone resorption on the bone slices. G–J – Immunoblotting study was done to study the levels of pAKT and NFATC1 in CSF-1 and ODF-induced bone marrow monocytes from wild type and OC-TG23b-3p mice; GAPDH was selected as a loading control
*P < 0.05, **p < 0.01 compared to wild type.
Discussion
In the present study, we evidenced that miR-23b-3p is over-expressed during macrophage colony stimulating factor and ODF-mediated osteoclastogenesis from bone marrow monocytes. We found that miR-23b-3p targets PTEN and activates the PI3/Akt/NFATC-1 cascade. In the in vivo studies, we found that the osteoclast-specific upregulation of miR-23b-3p suppresses both the bone volume and bone mineral density by enhancing the osteoclast activity. As a result of ODF treatment, the bone marrow monocytes from OC-TG23b-3p mice showed more substantial character to differentiate into osteoclasts. The protein levels of PTEN were suppressed, NFATC-1 and p-AKT were overexpressed, and in accordance with this the activity of osteoclast also increased; the findings were in accordance with the study which confirmed role of mir-23b-3P in pancreatic cancer through the Petn/Pi3K/Akt pathway [31]. To date, numbers of miRs have been identified to be associated with regulation of differentiation of osteoclasts [37–41]. MiR-148a and miR-21 have been found to induce osteoclastogenesis via targeting MAFB and PDCD-4 respectively. We came across a study involving patients with osteoporosis which suggested that the expression of about 754 different miRs was altered in osteoporosis; among them miR-23b-3p and miR-140-3p were significantly upregulated [23]. In the present study, we established that miR-23b-3p plays a potential role in osteoclastogenesis and alters the activity of osteoclasts via regulating the levels of osteoclast genes. PTEN is identified to be the favorable target for miR-23b-3p through 3′-UTR reporter assays [42]. The reports suggested that miR-23b-3p prevents multiple autoimmune diseases via regulation of inflammatory cytokine pathways [43]. In the present study, we demonstrated that miR-23b-3p promotes osteoclastogenesis via targeting PTEN and not STAT3 in ODF and CSF-1 induced TIB-71 cells, though both are necessary for functioning of osteoclasts. The results hence suggest the importance of miR-23b-3p in controlling the expression of genes associated with bone remodeling.
Osteoclast and osteoblast lineages play an important role in remodeling and maintaining mineralization of bones [44, 45]. MiRNAs are responsible for regulation of genes and hence coordinate a number of cellular processes involved in regulating bone remodeling. However, the involvement of miRs establishing a correlation between the two different cell types of opposing characters, i.e. osteoclasts and osteoblasts, remains poorly explored. The involvement of miR-23b-3p in the regulation of osteoclasts and osteoblasts implies that it could be involved in such a correlation between these two cell types. It has been evidenced previously that miR-23b-3p plays a prominent role in exosome-mediated microRNA signaling in breast cancer [46]. The results hence suggest involvement of a potential mechanism in which miR-23b-3p mediated a link between osteoclasts and osteoblasts.
Overall, in conclusion, the present study suggests that miR-23b-3p regulates differentiation of osteoclasts via targeting PTEN. The study indicates a novel role of miR-23b-3p in osteoclastogenesis. In this study, we highlighted the potential of miR-23b-3p in regulating bone loss via acting directly on osteoclasts. In conclusion, miR-23b-3p antagonist promotes bone marrow formation via blocking osteoclasts and causing promotion of osteoblasts simultaneously. Blocking the expression of miR-23b-3p could be a new therapeutic approach in controlling osteoporosis in elderly patients as well as in postmenopausal women. Further studies are needed to clarify the mechanisms involved in overexpression of miR-23b-3p in osteoclasts and the bridging role of miR-23b-3p in osteoclasts and osteoblasts.