Introduction
According to the American Cancer Society, there were over 135,000 new cases of colorectal cancer (CRC) (95,520 colon cancers and 39,910 rectal cancers) in 2017 in the United States. Due to the large number of new cases, colorectal cancer was in the third place among the cancerous causes of deaths in men and women, which resulted in more than 50,000 deaths for this reason in the US in 2017, and about 655,000 worldwide. The risk of this type of cancer is slightly higher in men than in women [1–3].
Many aspects are mentioned among the causes of development of CRC. First of all, scientists have acknowledged that the main factor which increases the risk of the disease is inheritance of mutations from first-degree relatives. Almost 30% of patients have at least one relative in the family who suffers from CRC [2]. The second factor that predisposes to CRC is familial adenomatous polyposis (FAP), which appears in around 1% of all CRC cases. Another disease which can predispose to CRC is chronic inflammatory bowel disease, as known as Crohn disease and ulcerative colitis. Authors, among risk factors, also mention diet and lifestyle, type 2 diabetes, cigarette smoking, alcohol abuse, obesity, diet low in fiber, and an excess of consumed fats, carbohydrates as well as red and processed meat. Physical activity and long-term treatment with low doses of aspirin may have preventive value in the development of CRC. These factors have different influence on particular parts of the colon, sigmoideum and rectum [1, 2].
The RAS and RAF family proteins mediate signaling of growth factor receptors via the PI3K-AKT-mTOR and RAS-RAF-MEK-ERK pathways, thereby participating in cell survival and proliferation [4]. Excessive activity of these signaling pathways is often found in various cancers. It is caused mainly by mutations in RAS and BRAF genes. Based on the deficiency of DNA repair and influence of carcinogens, these oncogenes are often mutated in CRC patients. Right-sided colon cancer is characteristic for women and probably shows microsatellite instability as well as BRAF mutations. Left-sided colon cancer is more common in men and shows chromosomal instability as well as KRAS mutations. Detection of abnormalities in the KRAS, NRAS and BRAF genes is extremely important for proper qualification of patients for panitumumab and cetuximab therapy, which have been authorized by the Food and Drug Administration (FDA) and European Medicines Agency (EMA) on the basis of several clinical trials, including PRIME (panitumumab) and CRYSTAL (cetuximab) studies [5–11].
In our study, we examined the frequency of mutations in the KRAS, NRAS and BRAF genes in a large group of Caucasian CRC patients. The molecular tests were performed during the routine diagnostic process in qualification of CRC patients for first line chemotherapy with anti-EGFR (epidermal growth factor receptor) antibodies. For the first time, we examined the relationship between the exact location of CRC and the presence of particular mutations.
Material and methods
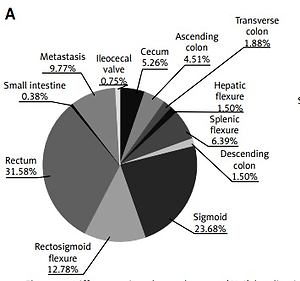
The study group included 500 patients (200 women and 300 men) with CRC including cancers in the small intestine (ICD-10: C17), colon (ICD-10: C18), rectosigmoid flexure (ICD-10: C19), rectum (ICD-10: C20) and anus (ICD-10: C21). The median age for men and women was the same: 66 years. 447 patients had locally advanced disease, while 53 patients had metastases at the time of diagnosis (with available material from the metastases). Patients were characterized in terms of age, gender and tumor localization. In the studied population, rectal and sigmoid cancers were the most common (61% of all CRC patients). Detailed characteristics of our group are presented in Table I.
Table I
Patient characteristics
DNA was isolated from formalin-fixed, paraffin-embedded (FFPE) tissues using the Qiagen QIAamp DNA FFPE-kit with the CE-IVD certificate. Tissue was collected and mutations were searched at the time of the diagnosis of colon and rectum cancer. The DNA was isolated from a paraffin block containing at least 50% of tumor cells. The percentage and presence of cancer cells were confirmed in the pathomorphological examination. FFPE samples were collected in 2012–2018. Analysis of mutations in the KRAS, NRAS and BRAF genes was carried out using three kits of the KRAS/BRAF, NRAS and BRAF Mutation Analysis Kit for Real-Time PCR (EntroGen, CE-IVD), on Cobas 480 real-time PCR apparatus (Roche Diagnostics). The tests examined the most common mutations in codons 12, 13, 59, 61, 117 and 146 in KRAS and NRAS genes, as well as in codon 600 of the BRAF gene. The tests can detect a mutation load of less than 1%. This sensitivity greatly depends on the extent of fragmentation and quality of the isolated DNA.
No attempt was made to find mutations in whole blood due to the availability of only FFPE tissues.
Statistical analysis
Statistical analysis was performed using the χ2 test to determine the relationship between different tumor localization and the occurrence of mutations. Results were statistically significant when the p value was below 0.05.
The study was approved by the Local Ethical Committee of the Medical University of Lublin (no. KE-0254/218/2015).
Results
Frequency of KRAS, NRAS and BRAF mutations in colorectal cancer
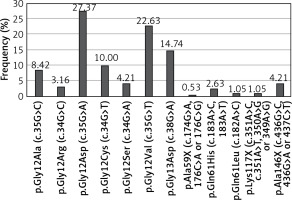
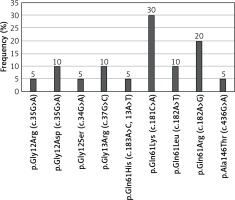
KRAS mutations were detected in 190 (38%) patients, NRAS mutations in 20 (4%) patients, and BRAF mutations in 24 (4.8%) patients. The most common substitution in the KRAS gene was p.Gly12Asp (27.37% of all KRAS mutations; 52/190). 90.53% of KRAS mutations occurred in codon 12 or 13 (Figure 1). The most common substitution in the NRAS gene was Gln61Lys (30% of all NRAS mutations; 6/20). 65% of NRAS mutations were found in codon 61 (Figure 2). Among all BRAF mutations, only Val600Glu was found.
Association between age, gender, tumor localization and mutation status
There were no associations between age of CRC patients and frequency of KRAS, NRAS and BRAF gene mutations. These mutations were significantly more often diagnosed in women (55.5% of female patients; 111/200) than in men (41% of male patients; 123/300, p < 0.005). The frequency of KRAS mutations and BRAF mutation was significantly higher in female than in male patients (χ2 = 8.266, p = 0.0044 and χ2 = 4.14, p = 0.042, respectively), while the frequency of NRAS mutations was similar in both sexes (χ2 = 0.1, p = 0.75) (Tables I and II).
Table II
Relationship between occurrence of mutations in the KRAS, NRAS and BRAF genes and sex, age, and tumor localization in colorectal cancer (CRC) patients
Rectal and sigmoid cancers were the most often diagnosed tumors in both groups of patients: with and without KRAS, NRAS or BRAF gene mutations (Table I). However, only 28% of patients with KRAS mutations (53/190) and up to 45% of patients with NRAS mutation (9/20) had rectal cancer. The most common tumor localization in patients with BRAF mutations was the cecum (21% of patients with this mutation; 5/24) (Table I). Mutations were most often found in tumors of the transverse colon (70.6% of all patients with transverse colon cancer, 12/17, p < 0.05 in comparison to other CRC localizations) and the ascending colon (73% of all patients with ascending colon cancer, 27/39, p < 0.005 in comparison to other CRC localizations) as well as in cecum cancer (62.2% of patients with cecum cancer, 23/37, p = 0.0516 in comparison to other CRC localizations). Patients with cancers of the sigmoideum (38.2% of all sigmoid cancer, 39/102, p = 0.052 in comparison to other CRC localizations) and splenic flexure (22.7% of all splenic flexure cancer, 5/22, p < 0.05 in comparison to other CRC localizations) had mutations confirmed the least frequently. Mutations were significantly more often found in patients with colon cancer including cancer of the cecum (56.7% of mutated tumors) than in patients with sigmoid and rectal cancers (42.6% of mutated tumors, p = 0.002, χ2 = 9.57) (Table II). The mutation significantly more frequently occurred on the right side of the large intestine (65% of this localizations of tumor, 63/97) than on the left side of the large intestine (40.8% of this localization of tumor, 141/366). The incidence of KRAS and BRAF genes varied depending on the CRC localization in the right or left parts of the large intestine. In contrast, mutations in the NRAS gene occurred at a similar frequency in these two CRC localizations (Table III).
Table III
Relationship between occurrence of mutations in the KRAS, NRAS and BRAF genes and tumor localization in colorectal cancer (CRC) patients. *Lack of any examined mutations (wild type)
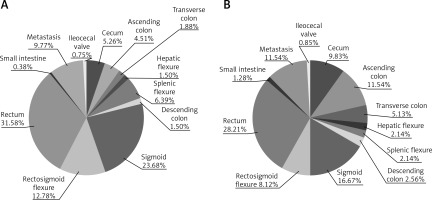
The differences in the occurrence of individual CRC in men and women with and without mutations in the KRAS, NRAS and BRAF genes were not significant. However, mutations in the KRAS gene in the small intestine and in the ileocecal valve were found only in female patients (Figure 3). Male patients with NRAS mutations suffered primarily from rectal cancer (57.1% of all men with NRAS mutations) and cancer of the rectosigmoid flexure (14.3% of all men with NRAS mutations). Single cases of sigmoid, ascending colon and small intestine cancers were found in men with NRAS mutations, whereas 50% of female patients with NRAS mutations suffered from colon cancer. Two cases of sigmoid and 1 case of rectal cancer were found in female patients with NRAS mutations. 45.5% of men and only 23.1% of women with BRAF mutation had rectal or sigmoid cancers, whereas colon (38.5% of all women with BRAF mutations) and cecum (23.1% of all women with BRAF mutations) cancers predominated in female patients with BRAF mutations.
Discussion
Frequency of KRAS, NRAS and BRAF gene mutations was assessed in previous clinical trials that evaluated the efficacy of anti-EGFR antibodies in the first and third line of treatment in CRC patients [4, 11–15].
The lack of efficacy of cetuximab combined with first line chemotherapy with 5-fluorouracil and oxaliplatin in patients with KRAS gene mutations was demonstrated in the OPUS study (the study group consisted of 314 patients). KRAS codons 12 and 13 mutations were found in 43.2% of CRC patients and the Val600Glu BRAF mutation in 3.5% of CRC patients. Efficacy of cetuximab was observed only in patients with the wild-type KRAS gene (codons 12 and 13). Rare RAS mutations were examined in archival material a few years later using the BEAMing technique. 26.3% of patients without KRAS codons 12 and 13 mutations had rare RAS mutations, including KRAS mutations in codon 59 or 61 (5.9% of patients), in codon 117 or 146 (9.3% of patients) as well as NRAS mutations in codon 12 or 13 (6.8% of patients), in codon 59 or 61 (in 5.1% of patients) and in codon 117 or 146 (0.8% of patients). Patients with rare RAS mutations also did not benefit from cetuximab therapy [15, 16].
The efficacy of first line chemotherapy based on irinotecan and 5-fluorouracil with or without cetuximab in patients without KRAS mutations (codons 12 and 13) was examined in the CRYSTAL study. KRAS gene mutations in codon 12 or 13 were found in 37.3% and the Val600Glu mutation in the BRAF gene in 6.6% of CRC patients (the study group consisted of 1063 patients). The benefit of cetuximab was not the same in all patients with the wild-type KRAS gene (codons 12 and 13). Therefore, rare RAS mutations in patients enrolled in thee CRYSTAL trial were examined. 14.7% of 430 patients with wild-type KRAS codons 12 and 13 had rare RAS gene mutations. Mutations in codons 59 and 61 of the KRAS gene were present in 3.3% of patients and mutations in codons 117 and 146 were present in 5.6% of patients. NRAS gene mutations were found in codons 12 and 13 in 3.5% of patients, in codons 59 and 61 in 2.8% of patients, and in codons 117 and 146 in 0.9% of patients. Effectiveness of cetuximab in patients with rare RAS mutations was unsatisfactory [9, 17].
The PRIME clinical trial compared the efficacy and safety of panitumumab, 5-fluorouracil and oxaliplatin with chemotherapy alone in the first-line treatment of 1096 CRC patients. 67% of patients had KRAS codon 12 or 13 mutations. KRAS codon 61 was mutated in 4% of patients and codon 117 or 146 was mutated in 6% of patients. Mutations in the NRAS gene in codons 12 and 13 were found in 3% of patients and in codon 61 in 4% of patients. There were no mutations in codon 117 or 146 of the NRAS gene. Mutations in the BRAF gene occurred in 8% of CRC patients. The effectiveness of panitumumab was closely related to the absence of mutations in RAS and BRAF genes [8].
In the PEAK study (221 patients with known status of examined genes), the effectiveness of panitumumab monotherapy in third line treatment of CRC patients was examined. The following mutations were found in CRC patients from the PEAK trial: in codon 12 or 13 of the KRAS gene in 43.1% of patients, in codon 59 or 61 of the KRAS gene in 4.8% of patients, in codon 117 or 146 of the KRAS gene in 5% of patients, in codon 12 or 13 of the NRAS gene in 4.2% of patients, in codon 59 or 61 of the NRAS gene in 3% of patients and in codons 117 and 146 of the NRAS gene in 1.1% of patients. The occurrence of the mutations was closely related to the lack of efficacy of panitumumab [18].
The incidence of examined mutations in our patients is lower than in the cited studies. Mainly, the frequency of mutations in the KRAS and BRAF genes is lower than in the CRYSTAL and PRIME studies. This is probably due to the lower sensitivity of real-time PCR technique used in the routine diagnosis of RAS and BRAF mutations in CRC patients in our study. The results of our study indicated that the most frequent mutation in the KRAS gene was Gly12Asp and in the BRAF gene was Val600Glu, according to the results of genetic tests carried out in clinical trials. Mutation in codon 61 was the most frequent mutation in the NRAS gene in our patients, which does not match with the results of other studies. The research should be supplemented by demonstrating that tumor heterogeneity and/or low sensitivity of diagnostic tests may have contributed to the fact that patients with wild-type RAS did not respond to anti-EGFR therapy due to the presence of mutations. The weakness of our study was the lack of information on how the patients were treated.
Kodaz et al. studied the relationship between the prevalence of KRAS mutations and the clinicopathological characteristics of colorectal cancer. The study group included 189 patients with CRC diagnosis. 47.6% of patients had a mutation in the KRAS gene. The study also showed that the most common KRAS mutations occurred in codon 12 (73.3% of all KRAS examined mutations) and the most common substitution was Gly12Asp (42.4% of all KRAS examined mutations). The authors found that a high percentage of young CRC patients (< 40 years) had the wild-type KRAS gene. They also suggested that KRAS point mutations in colorectal cancer exhibited a heterogeneous distribution in terms of tumor localization. In the cited study, there was no significant difference in KRAS mutation frequency according to tumor localization. Moreover, the authors found no association between KRAS mutation occurrence and gender [19].
Kawazoe et al. searched for KRAS, NRAS, BRAF and PIK3CA gene mutations in the material from 246 patients with metastatic CRC. Fifty percent of patients had wild-type examined genes. Mutations in codons 12 and 13 of KRAS gene were found in 34.1% of patients, while mutations in codons 61 and 146 were detected in 10 cases (3.8%). NRAS gene mutations occurred in 11 patients (4.2%) and Val600Glu mutation in the BRAF gene occurred in 14 (5.4%) people. The authors stated that primary rectal tumors tended to be more frequently RAS-mutated and BRAF mutant tumors were more likely to develop in the right colon. They observed no significant association between RAS gene status and other clinicopathological features such as age, sex, primary lesion localization, histology, or site of metastases, which is similar to the results of the study by Morris et al. [20, 21].
In our results, mutations were found to be associated with sex and anatomical location of the tumor. We observed the highest percentage of tumors with KRAS, NRAS and BRAF gene mutations in colon cancers. Moreover, the highest percentage of tumors with mutations were found in the right side of the large intestine. A higher percentage of female patients had KRAS, NRAS or BRAF mutations than male patients, which was also observed by Ng et al. in their research [22].
KRAS mutations were reported to be more frequent in right colon tumors by Bleeker et al. [23] and Loree et al. [24], but in left colon tumors by Zulhabri et al. [25]. Watanabe et al. [26] and Sinicrope et al. [27] found that KRAS codon 12 or 13 mutations were significantly more frequent in the right colon. Yamauchi et al. [28] reported that KRAS mutations were more common in cecum tumors. Brink et al. [29] reported that KRAS codon 13 mutation was more common among females with rectal tumors.
Moretto et al. [11] conducted a study of 75 CRC patients with wild-type RAS and BRAF genes. They found that patients with tumors located on the right side of the large intestine more often did not respond to therapy based on cetuximab or panitumumab compared to patients with tumors on the left side of the large intestine. If we assume that mutations occur more often on the right side of the large intestine, this relationship may result from the problems with detection of RAS and BRAF gene mutations due to tumor heterogeneity or low sensitivity of molecular tests.
In conclusion, our study showed that the occurrence of mutations in the KRAS, NRAS and BRAF genes is not accidental and depends on the location of CRC tumors. In case of failure of treatment with anti-EGFR antibodies in patients with tumor localization suggesting a higher probability of mutation presence, an insightful molecular examination is necessary.