Birth defects account for a high proportion of infant mortality and morbidity in China [1]. With the increased average age of pregnant women, neonatal birth defects have become increasingly prevalent. Therefore, it is of great significance to carry out a prenatal diagnosis to reduce the birth rate of defective newborns. Chromosomal abnormalities are an important cause of birth defects; thus using different detection methods to detect and study chromosomal abnormalities is important and significant.
Multiple detection methods have been developed to detect chromosomal abnormalities in prenatal diagnosis, including chromosome karyotyping, fluorescence in situ hybridization (FISH), bacterial artificial chromosomes (BACs)-on-Beads (BoBs), chromosomal microarray analysis (CMA), and quantitative fluorescent polymerase chain reaction (QF-PCR) [2]. Although chromosome karyotyping can accurately detect the number and structure of chromosomes in amniotic fluid cells, it cannot detect chromosome microdeletion and microduplication. Additionally, the period of amniotic fluid cell culture is relatively long (at least 7 days), suggesting that the karyotyping method is time-consuming and not suitable for rapid diagnosis. To date, more than 300 microdeletion and microduplication syndromes have been reported worldwide, with the incidence rates ranging from 1/4000 to 1/50 000 [3]. Therefore, the importance of chromosome microdeletion and microduplication detection should be emphasized.
FISH can quickly detect trisomy 13, trisomy 18, trisomy 21, and numerical abnormalities of sex chromosomes; however, it requires specific probes to detect the microdeletion and microduplication of uncommon chromosomes. Instead, CMA can detect the chromosome deletion and duplication of the whole genome, however, the complicated results from CMA require plenty of time for analysis. In addition, CMA as a diagnostic tool also creates uncertainties when copy number variation (CNV) of unknown pathogenic significance (variant of unknown significance – VOUS) is detected and the test is relatively costly [4]. Recent studies have reported that BoBs technique has been used for the detection of aneuploidies, including trisomy 13, trisomy 18, trisomy 21, numerical abnormalities of sex chromosomes, as well as at least 9 microdeletion regions [5]. The BoBs technology is a cytogenetic assay for rapid prenatal diagnosis [6], and is a molecular detection technique based on the principle of liquid-phase gene microarray with biotin-labeled samples, hybridized with reference DNA and solidified into fluorescence-encoded microspheres, which are detected by specific BACs DNA probes [7]. The BoBs assay is a bead-based multiplex assay, based on PerkinElmer’s BoBs and Luminex’s technologies [5, 8]. Therefore, it could be used to detect multiple syndromes caused by these microdeletion regions, which include Angelman syndrome (AS), Prader-Willi syndrome (PWS), DiGeorge syndrome (DGS), Miller-Dieker syndrome (MDS), cri-du-chat syndrome (CdCS), Wolf-Hirschhorn syndrome (WHS), Smith-Magenis syndrome (SMS), Langer-Giedion syndrome (LGS), and Williams-Beuren syndrome (WBS) [9].
Here, a total of 2218 pregnant women with high-risk factors were enrolled in this study for evaluation of BoBs technique in clinical settings. Amniotic fluid cells were examined by combining BoBs technique with chromosome karyotyping, aiming to explore potential benefits of combination of the two testing methods in prenatal diagnosis. Furthermore, CMA was employed on patients whose cells have microdeletion/microduplication detected by BoBs technique with a single probe.
Methods
Subjects
A total of 2266 pregnant women with high-risk factors for chromosomal abnormalities admitted to Weihai Maternity and Child Care Hospital (Weihai, Shandong, China) in the period from September 2020 to December 2021 were enrolled in this study. The indications for prenatal genetic evaluation included advanced maternal age (> 35 years), high-risk non-invasive prenatal testing (NIPT) results, abnormal ultrasound results, high-risk serological screening results, chromosomal disorder history from the couples, and adverse pregnancy history. In this study, the pregnant women or their family members signed informed consent.
Specimen collection
During amniocentesis guided by B-ultrasound, 25 ml of amniotic fluid was extracted, 20 ml of which was used for the amniotic fluid cell culture and 5 ml of which was used for the BoBs detection.
Conventional karyotype analysis
Approximately 20 ml of amniotic fluid was centrifuged at 1,500 rpm for 10 min at 4°C to separate amniotic cells. The cells were cultured, harvested and mounted on glass slides for the chromosome G-banded analysis by the Leica system. On each slide, 20 stained metaphases were randomly selected and examined, and 6 karyograms were created for the chromosome analysis. If a suspicious chromosomal abnormality or a chromosomal polymorphism was found, the count number of metaphases was then increased to a number between 50 and 100 for a more reliable result.
BoBs assay
The DNA samples were extracted from approximately 5 ml of amniotic fluid using DNA extraction reagents according to the kit’s handbook (QIAGEN, Germany). Approximately 5 ml of amniotic fluid was centrifuged and the flow-through was discarded. 200 μl of Buffer AL was added, and mixed thoroughly by vortexing. It was incubated at 56°C for 30 min. We briefly centrifuged the 1.5 ml microcentrifuge tube to remove drops from the lid, added 200 μl of ethanol (100%), mixed it thoroughly by vortexing, briefly centrifuged the tube to remove drops from the lid, pipetted the mixture onto the QIAamp Mini spin column (in a 2 ml collection tube) and centrifuged it at 12 000 rpm for 1 min. Then we discarded the flow-through and collection tube, placed the QIAamp Mini spin column in a new 2 ml collection tube and added 500 μl Buffer AW1, centrifuged and discarded the flow-through, added 500 μl of Buffer AW2, then centrifuged and discarded the flow-through, placed the column in a new collection tube and centrifuged at full speed for 1 min, placed the column in a new 1.5 ml microcentrifuge tube, added 40 μl of AE and incubated it at room temperature (15–25°C) for 3 min, and centrifuged it at 12 000 rpm for 2 min to elute the DNA.
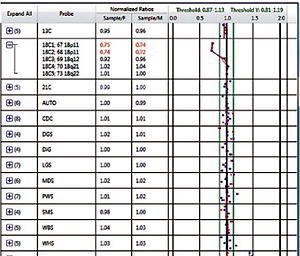
Male and female reference samples were used for the internal quality control standards and the comparison standards. A BoBs kit (Perkin Elmer, Waltham, MA, USA) was used for BoBs assay. The beads were analyzed using a Luminex 200 cytometric acquisition system (Austin, TX, USA) for data collection. Data were analyzed using BoBsoft 1.0 software. Results’ evaluation criteria: The normal ratio was 1.0. If the ratio was less than 0.8, it indicated that the probe was missing. If the ratio was more than 1.2, it indicated that the probe was repeated. The result suggested a deletion or duplication in the region if there were 3 or more probes in a certain target area exceeding the cut-off value of male and female reference, which indicated that the region was a deletion or duplication.
CMA assay
10 ml of amniotic fluid was extracted for the CMA analysis. Using Affymetrix’s CytoScan 750K chip, the amplification, hybridization, scanning and analysis were followed as the standard operating procedures of the Infinium HD Assay. ClinGen, ClinVar, DGV, OMIM, DECIPHER and other databases were used for interpretation of results. According to the classification standards recommended by the ACMG (PMID: 31690835), five categories were divided for the clinical determination: pathogenic, likely pathogenic, uncertain significance, likely benign, and benign.
Ethics approval and consent to participate
The authors declare that the experiments of the present study comply with the current laws of China. All patients signed written informed consent forms and agreed to the use of relevant data and information for scientific research. This study was approved by the ethics committee of Weihai Maternity and Child Care Hospital (WHFY-YXLL-WYH-L2022004).
Results
Cell culture results
The amniotic fluid cells from a total of 2218 cases were cultured for detection. The length of harvest time was related to the number and growth state of cells. In general, the average harvest time was 9 days, with the shortest harvest time of 8 days and the longest harvest time of 15 days.
Chromosomal results detected by BoBs technique
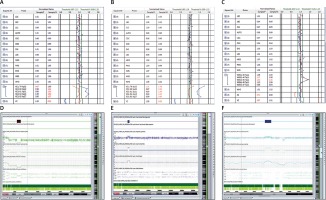
Out of the 2218 amniotic fluid samples, 74 cases were reported with chromosomal abnormalities, with a detection rate of 3.34% (Tables I and II). Among the 74 cases, 47 cases had numerical abnormalities at chromosomes 13, 18, 21, X and Y, which was consistent with the karyotyping results; 4 cases showed microdeletion at 5p15.3-15.2, 18p11.32p11.21, 22q11.2, and Y microdeletion, respectively; 1 case showed microduplication at 18p11.32p11.21 (Table I and Figure 1); 3 cases were reported with a chromosome missing and 9 cases were reported with chromosome duplication with the first probe of Xp22.31; 4 cases were reported with chromosome duplication with the probes of 7q11.23, including 1 case with the first probe, 1 case with the second probe, 1 case with the second and third probe, and 1 case with the third probe; 1 case was reported with chromosome duplication with the fifth probe of 8q24.11; 1 case was reported with chromosome duplication with the second probe of 22q11.21; 2 cases were reported with chromosome duplication with the fourth probe of 22q11.21 (Table II). The patients whose samples were reported with chromosomal abnormalities with the single probe detection were recommended for further CMA detection. Although 6 patients did not perform CMA, the CMA results of the remaining patients confirmed the chromosomal microdeletion/microduplication results reported by BOBs technique (Table II and Figure 2).
Table I
Comparison of BoBs and chromosome karyotype analysis
Table II
Microdeletions/microduplications detected by BoBs and CMA assay
Figure 1
Microdeletions/microduplications detected by BoBs assay. A – Microdeletion of 18p11.32p11.21; B – Microduplication of 18p11.32p11.21; C – Microdeletion of Y

Figure 2
Single probe deletion/duplication detected by BoBs assay and confirmed by CMA. A, D – BoBs assay results revealed microdeletion on the first probe of Xp22.31; SNP array verification was consistent with BoBs assay; B, E – BoBs assay results revealed microduplication on the first probe of Xp22.31; SNP array verification was consistent with BoBs assay; C, F – BoBs assay results revealed microduplication on the third probe of 7q11.23; SNP array verification was consistent with BoBs assay
Chromosomal results detected by karyotyping
We detected 95 cases of abnormal chromosomes with the detection rate of 4.28% (95/2218) by karyotyping. Among the 95 cases, 50 cases had a numerical abnormality of chromosomes and 45 cases had a structural abnormality of chromosomes (Table III). For the numerical abnormality, 11 cases were reported as chimera with the karyotyping results of 45, X0[11]/46, XX[39], 45, X0[5]/46, XX[55] and 47, XX, +mar[51]/46, and XX[9]. However, their corresponding results from BoBs technique were reported as being normal. The cases detected as the chromosome structure abnormalities (chromosomal translocation and inversions) by karyotyping were also detected as being normal by BoBs technique.
Discussion
Birth defects are congenital disorders caused by various abnormalities during pregnancy, which are the main reasons for the disability and even death of the newborns. It is well known that chromosomal abnormality is one of the causes of birth defects. In recent years, although the three-level prevention and control policy has been implemented to prevent and treat birth defects in China, there are still about 900,000 newborns with birth defects each year. Therefore, there is an urgent need to employ pre-pregnancy examination and novel neonatal disease screening technologies to prevent disability and health hazards of newborns [10].
Karyotyping is a gold standard test for detecting chromosomal aberrations in prenatal diagnosis. However, it has several drawbacks, including 1) the long cell culture time (at least 7 days) of the amniotic fluid cells; 2) the potential contamination during the cell culture; 3) few cells at the division phase; 4) the higher failure rate of the amniotic fluid cell culture with the increase of gestational ages; 5) the low loci resolution (greater than 10 Mb in size). Since September 2020, we have introduced the BoBs technique to combine with karyotyping for a more rapid and comprehensive prenatal diagnosis, with additional detection of aneuploidies at 9 microdeletion/microduplication regions.
In our study, the numerical abnormalities at chromosome 13, 18, 21, X, and Y detected by BoBs were found to be consistent with the results from the karyotyping analysis. Briefly, 2 cases of trisomy 13 were detected by BoBs, with the corresponding karyotyping results of 47, XN, +13. Six cases of trisomy 18 were detected by BoBs, five of which had the corresponding karyotyping result of 47, XN, +18 and one of which had the corresponding karyotyping result of 47, XY, +18[15]/46, XY[45]. One case of microduplication at 18p11.32p11.21 was detected by BoBs, with the karyotyping result of 46, XN, -21, +der(18)t(18; 21)(q11.2; q11.2), and its corresponding CMA result of 18p11.32p11.21(136,228-15,079,294) x1, 18q22.3q23(68,960,865-78,013,728)x1. 32 cases of trisomy 21 were detected by BoBs, with thirty corresponding karyotyping results of 47, XN, +21, one corresponding karyotyping result of 47, XX, +21[55]/46, XX[5], and one corresponding karyotyping result of 47, XX, +21[33]/46, XX[27]. In addition, 6 cases were detected as sex chromosome abnormalities by BoBs technique, of which two had karyotyping results of 45, X0, one of 45, X0[30]/46, XY[20], one of 47, XXY[68]/48, XXYY[17]/49, XXYYY[15], one of 45, X0[38]/46, X, der(X)t(X; 4)(q28;q26)[12], one of 47, XXY, and one of 47, XXX. In contrast, 2 cases with normal results from BoBs technique were identified as chromosome abnormalities by karyotyping, with results of 45, X0[11]/46, XX[39] and 45, X0[5]/46, XX[55], respectively. Previous studies indicated that the detection threshold of BoBs on chimera was 20–40% [11]. Our results here also indicated the limitation of BoBs technique in the detection of a low proportion of chimeras. Additionally, the results of 42 amniotic fluid samples from karyotyping were reported as being abnormal, while their corresponding results detected by BoBs technique were shown as being normal, indicating that BoBs technique was incapable of detecting the balanced translocation of the chromosomes.
In addition to its rapid detection of aneuploidy, BoBs technique could also be used to detect chromosomal microdeletion and microduplication efficiently and sensitively. Our BoBs results identified 2 cases of microdeletion syndromes, including 5p15.3-15.2 microdeletion syndrome and 22q11.21 microdeletion syndrome. Therefore, the couples were recommended for genetic counseling and further chromosomal testing of maternal blood. It would be significant for their second conception if one of the couples were a carrier or chimera [12, 13].
Moreover, we identified 22 microdeletion/microduplication cases by BoBs technique with a single probe. Briefly, 12 microdeletion/microduplication events occurred at the Xp22.31 region with the variation rate of 0.54% (12/2218) (Table II, Figures 2 A and B). Previous studies indicated that steroid sulfatase (STS) was located in the Xp22.31 region. It has been reported that the deletion of the STS gene was associated with the X-linked recessive ichthyosis disease (XLRI) [14, 15]. Moreover, repetition of the Xp22.31 fragment was associated with developmental delay, autism, language development delay, hypotonia, epilepsy, congenital finger flexion, precocious puberty, hyperopia, digital flexion, absence seizures, and other clinical phenotypes [16, 17].
In addition, 4 cases were detected as microduplication at the 7q11.23 region by BoBs technique, one of which underwent further CMA with the result of 7q11.23 (72,723,371-74,136,633)x3 (Table II and Figures 2 C and F). The 7q11.23 region contains the elastin (ELN) gene. It has been reported that duplication of the ELN gene in the 7q11.23 region was associated with the autosomal dominant 7q11.23 duplication syndrome [18]. The 7q11.23 duplication syndrome is a rare chromosomal microduplication syndrome, with a prevalence of approximately 1/12,000, the clinical phenotypes of which include facial abnormalities, cardiovascular disease, neurological abnormalities, speech disorders, behavioral abnormalities, developmental delays, and intellectual disabilities [19, 20]. One case was identified as microduplication by the fifth probe at the 8q24.11 region with its further CMA result of 8q24.11 (118,225,217-118,884,779)x3, confirming our BoBs analysis. However, the potential impact of duplication of the 8q24.11 fragment was still unclear (Table II).
Moreover, 2 cases were identified as microduplication at the 15q11.2q13.1 region by the third probe with their CMA results of 15q11.2q13.1 (22,770,422- 28,397,034)x3 and 15q11.2q13.1 (22,770,422-28,540,345)x3 respectively, further validating our BoBs results (Table II). Angelman syndrome and Prader-Willi syndrome were caused by dysfunctional genes located in the 15q11.2q13.1 region. The region contains 24 OMIM genes which were associated with genetic imprinting. Duplication of the 15q11.2q13.1 region would cause various clinical phenotypes [21]. Therefore, we recommended further genetic counseling and CMA on parental amniotic fluid specimens for examining the origin of the 15q11.2q13.1 duplicated region with parental amniotic fluid specimens. One case was identified as microduplication at the 22q11.21 region by the second probe and two cases by the fourth probe. The 22q11.21 region contained 49 OMIM genes, including CLTCL1, HIRA and TBX1 (Table II). Duplication of this region could lead to mental retardation, learning disabilities, growth retardation, and dystonia [22, 23].
In conclusion, our study demonstrated that the combination of traditional karyotyping and BoBs technique is essential for the rapid and accurate prenatal diagnosis and provides necessary information for genetic counseling and further examinations. BoBs technique could accelerate the speed of prenatal diagnosis, increase the detection rate, and reduce the incidence of birth defects. Although the microdeletion/microduplication results detected by BoBs techniques with a single probe were not included in the final diagnostic report, we confirmed all these results with CMA, indicating the accuracy of BoBs technique. We believe that this information is beneficial to the patients if they can be included in the diagnostic report in the future. Additionally, based on the massive results about the functional knowledge of key genes in these microdeletion/microduplication regions in the final diagnostic report, we believe our study will provide more comprehensive guidance for prenatal diagnosis.