Introduction
Helicobacter pylori is a well-known causative agent of chronic dyspepsia, peptic ulcer, gastric mucosa-associated lymphoma and gastric cancer [1–3]. Currently, the global infection rate of H. pylori exceeds 50% of the population [4]. The treatment of H. pylori mainly comprises triple therapy such as omeprazole, clarithromycin and amoxicillin [5]. However, recent studies show that the curative rate of the triple therapy is gradually decreasing, mainly due to the emergence of resistant strains [6, 7]. Recently, the WHO has considered clarithromycin-resistant H. pylori to be one of the most urgently needed new antibiotic groups [8]. The mechanisms of resistance to clarithromycin mainly result from mutations of the peptidyl transferase region encoded in domain V of 23s rRNA [9]. The increase in clarithromycin resistance in the last decade has reduced the efficacy to eradicate H. pylori [10].
In the past decade, various antibacterial peptides were discovered through the studies of organisms’ immune mechanisms [11]. These peptides showed antibacterial effects towards various kinds of bacteria, including H. pylori. For example, Chen et al. identified the first anti-H. pylori peptides from amphibian skins [12]. The peptide was composed of 23 amino acids with the sequence of GLLRASSVWGRKYYVDLAGCAKA. Another recent example is the venom peptide bicarinalin, which has produced similar anti-H. pylori activity as four antibiotics currently used in therapies against H. pylori [13]. Most of the discovered antibacterial peptides have a nonspecific innate immune response against exogenous pathogens. At present, the mechanisms of antibacterial activities can be divided into two groups. The first is the amphipathic α-helix structure of peptides that binds to the bacterial cell membrane to form pores and destroy the cell membrane by releasing the cell contents [14]. The second is the antibacterial peptides that directly enter the bacterial cells and act on the DNA, RNA, protein, mitochondria and cell membrane, inhibiting the synthesis, transcription and translation of the cells. The peptides may also interfere with cell metabolism and inhibit the formation of the cell membrane [14]. The study also demonstrated that peptides can kill bacteria at a faster rate than that of its multiplication [15]. The combination of the physical killing mechanisms and fast killing rate means resistant strains are potentially less likely to develop compared to most antibiotics [16]. Another advantage of antibacterial peptides is the highly degradable properties, which may lead to few drug residues and side effects [17].
Antibacterial peptides have been found in mammals, plants, insects and even microorganisms. This inspired us to evaluate whether food also contains anti-H. pylori peptides. The aim of this study was to investigate whether the most common food source, milk, contains any peptides against H. pylori. Milk is rich in protein, of which caseins account for about 80% of the total protein [18]. Caseins, in addition to nutrition, produce numerous biologically active peptides through enzymolysis during food processing. Many of these peptides have demonstrated antimicrobial activities. For example, bovine αs1-casein f (99–109) has antibacterial activities against Gram-positive bacteria Bacillus subtilis and Listeria innocua [19] and αs2-casein f (181–207), f (175–207) and f (164–207) demonstrated antibacterial properties against both certain Gram-positive and Gram-negative bacteria [19].
This study aimed to extract antibacterial peptides from caseins and to determine their minimum inhibitory concentrations (MIC) against H. pylori under environments with different pH values.
Material and methods
Caseins (Sigma C5890) from bovine milk were purchased from Sigma (Guangzhou, China). Antibacterial peptide extraction was performed on the caseins, followed by concentration and identification of the peptides. A study had shown that casein has anti-E. coli activities, and hence it was used in this study to ensure the extracted antibacterial peptides were active. The antibacterial activities of the extracts were also tested on H. pylori (ATCC-43504). Eight concentrations of the extracts were used to calculate the MIC values at four environments with different pH values. The eight concentrations were 25 mg/ml, 12.5 mg/ml, 6.25 mg/ml, 3.125 mg/ml, 1.5625 mg/ml, 0.78125 mg/ml, 0.390625 mg/ml and 0.1953125 mg/ml. They were all tested in environments with pH values of 1.5, 2.0, 2.5 and 3.0. The H. pylori were purchased from Tin Hang Technology, Hong Kong, China. The experiments were performed with blank solutions, negative and positive controls. The blank solutions did not contain antimicrobial peptides or H. pylori. They were used to ensure there was no contamination and also for measurement of the microplate spectrophotometer optical density values at 600 nm (Multiskan GO, Thermo Fisher Scientific). The negative controls contained H. pylori and no antibacterial peptides, whereas the positive controls contained the extracted antibacterial peptides and no H. pylori.
Recovery, storage, and identification of H. pylori
Strains of H. pylori (ATCC-43504) were recovered in accordance with the instructions provided by the supplier product information sheet (https://www.atcc.org/~/ps/43504.ashx). The cryo-preserved strains were thawed in a 37°C water bath. The solutions (100 μl) were then inoculated with 6% sheep blood Columbia agar (Huankai Microbial, Guangdong, China) and incubated at 37°C for 3 days in a facultative anaerobic environment. In terms of storage, glycerin was added to brain heart infusion broth to prepare the cryopreservation medium, where the H. pylori were added and stored at –80°C. Identification of the H. pylori (ATCC-43504) was performed by assessment of colony morphology by microscopy, in which conformation was suggested to be needle-like with a glassy translucent appearance of colonies formed on the Columbia blood agar plate (Huankai Microbial, Guangdong, China). This indicates that the strains were Gram-negative spiral bacilli. Identification of H. pylori (ATCC-43504) was also performed by biochemical assays, including oxidase, catalase and urease tests (Huankai Microbial, Guangdong, China). A combination of the above results served as the identification of the H. pylori strain.
Extraction and identification of antibacterial peptides
Caseins (20 g) were dissolved in 300 ml of deionized water at 37°C; the pH value of the solutions was adjusted to 1.5, 2.0, 2.5, or 3.0 with hydrochloric acid. Pepsin (0.6 g) (1 : 10000, P-5144, BoMei Biotechnology, HeFei, China) was added to the solutions for enzymolysis, which was carried out at 37°C for 5 h. During the experiments, hydrochloric acid was added drop-wise to maintain the pH value of the solution. After enzymolysis, the pepsins were inactivated by incubation in a water bath at 80°C for 20 min. The de-enzymatic solutions were then centrifuged at 3500 rpm and 9000 rpm for 30 min and 20 min, respectively. The supernatants were then collected and filtered through a 0.45 μm filter and re-filtered through a 0.22 μm filter. The mixtures were evaporated to dryness on a rotary evaporator and freeze-dried. The dry mixture (0.1 g) was sent to the National Center for Protein Sciences (Beijing, China) for molecular weight analysis and peptide sequencing analysis by MALDI-TOF mass spectrometry and MS/MS Ion Search, respectively.
Preparation of H. pylori and E. coli 0.5 McFarland Turbidity solution
Several H. pylori colonies from the 6% sheep blood Columbia broth were added to sterile tubes containing 0.9% normal saline, shaken for 15 s and adjusted to a turbidity of 0.5 in the microbial turbidimeter (DensiCHEK Plus, bioMerieux, USA). The same methods were performed for the preparation of the E. coli (ATCC-25922) solution.
Antibacterial activity test
Microdilution methods were used to evaluate the inhibitory effects of each group of antibacterial peptides against E. coli. Sterile Brucella broth (HopeBio, Qingdao, China) was used to dilute the antibacterial peptide extracts with pH = 1.5, 2.0, 2.5 and 3.0 through the following methods. The broth (600 μl) was added to a 48-well plate and 600 μl of 50 mg/ml antibacterial peptides was added to the first well, followed by seven half-dilutions to produce the 25 mg/ml, 12.5 mg/ml, 6.25 mg/ml, 3.125 mg/ml, 1.5625 mg/ml, 0.78125 mg/ml, 0.390625 mg/ml and 0.1953125 mg/ml peptide solutions. The E. coli 0.5 (approximately 1.5 × 108 CFU/ml) McFarland Turbidity solution (2 μl) was then added to each of the peptide solutions and incubated at 37°C for 24 h in a facultative anaerobic environment. The mixtures were assigned to five categories by naked-eye observations. The categories were clearly transparent, blurred (80% suppressed), turbidity significantly reduced (50% inhibited), turbidity mildly reduced and turbidity was not reduced. Mixtures of blurred or turbidity significantly reduced categories were considered of having antibacterial activities against E. coli.
Minimum inhibitory concentration determination
The preparations of MIC assays of antibacterial peptides against H. pylori were the same as those for analysis of E. coli, with the difference being that each H. pylori assay was repeated 12 times for higher accuracy and consistency. Instead of performing naked-eye observation as in the E. coli assays, the H. pylori MIC assays were performed in sterile 96-well microliter plates (ChunBo Biologics, Haimen, China) and the absorbance of the spectrophotometer was set at 600 nm. The MIC90 values were defined as the lowest concentration of peptide that inhibits 90% of H. pylori. The inhibition percentage was calculated by the following equation: Inhibition (%) = (1 – (Optical density (OD) value of sample/Optical density (OD) value of negative control)) × 100%.
Statistical analysis
Data analysis was performed using the paired t-test comparative method embedded in the SPSS 20.0 software to calculate the p-values, which were considered to be statistically significant when p was less than 0.05. The sample groups were compared with the blank control, negative and positive control groups. Regression analysis was used to analyze the relationship between the concentration of each group of the antibacterial peptides and their OD values. Pearson correlation analysis was used to evaluate the relationship between pH changes and MIC values.
Results
Antibacterial peptide extraction rate
Casein (20 g) was hydrolyzed by pepsin at different pH values. It was then concentrated and freeze-dried to give the weight and extraction rate as shown in Table I.
Identification of antibacterial peptides
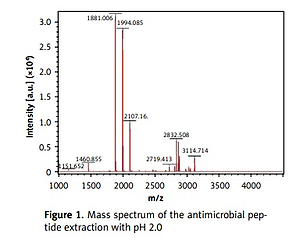
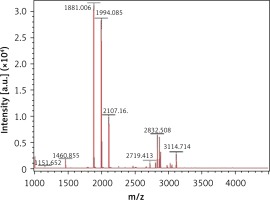
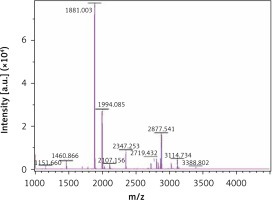
The MALDI-TOF mass spectrometry analyzed the molecular weight of the antimicrobial peptides at pH 2.0 and pH 3.0 (Figures 1 and 2). The peaks with the highest abundance at both pH values were located at 1881 m/z and 1994 m/z. Peptide sequencing analysis was performed by MS/MS Ion Search and the resulting amino acid sequences with 1881 m/z and 1994 m/z were YQEPVLGPVRGPFPIIV and LYQEPVLGPVRGPFPIIV, corresponding to residues 208–224 and 207–224, respectively (Table II). These sequences were matched with the corresponding peptides in the Antimicrobial Peptide Database [20], Milk Peptide Database [21] and MilkAMP Database [22]. The peptides with 1881 m/z were identified as casecidin 17 with APD (Antimicrobial Peptide Database) ID of AP01398, and there were no records in the above three databases for the peptide with 1994 m/z. Literature searches of this peptide were performed on the most common databases, including PubMed, Google Scholar, Cochrane Library Databases, and Science Direct, and only a single paper was found that mentioned its bioactivity, which is modulation of the bitter taste receptors and was named β-casein 207–224 [23].
Table II
Peptide profiles of the enzymolyzed casein at pH 2.0 and 3.0
Antibacterial activity and minimum inhibitory concentration determination
Antibacterial assays of the peptides against E. coli were performed using naked-eye observations. All the mixtures were assigned as blurred or turbidity significantly reduced, which indicate that E. coli were successfully inhibited by each group of antimicrobial peptides. This proved that the extracted antimicrobial peptides in each group were active.
With regards to the H. pylori MIC assays, all eight concentrations of the antibacterial peptides showed anti-H. pylori activity. The MIC90 of both pH 1.5 and pH 2.0 was 6.25 mg/ml, whereas for both pH 2.5 and pH 3.0, the MIC90 was 12.5 mg/ml.
Regression analysis showed that when the concentration of the peptide increased, the optical density (OD) values became smaller (p < 0.001) (Table III), indicating that the higher the concentration of the peptide is, the better is the anti-H. pylori effect. The Pearson correlation analysis between pH and the MICs of the antibacterial peptides in different concentrations showed a positive correlation between the pH and the MIC value with a correlation coefficient of 0.82. This indicates that the greater the pH value is, the larger is the MIC90 (p < 0.001). Hence, the anti-H. pylori effects of the peptides are higher in a lower pH environment, such as in the human stomach.
Table III
Optical density values of the extract with different concentrations and at environments with different pHs
Discussion
Due to an increase in H. pylori resistance to antibiotics, such as clarithromycin, the triple therapy treatment has become gradually less efficacious [8]. This study discovered that the major products of bovine milk casein digestion have promising anti-H. pylori effects. The two major peptides identified in this study were casecidin 17 and β-casein 207–224. The anti-H. pylori effects of bovine milk have been suggested by Wang et al. [24] and the non-H. pylori specific antibacterial activities against both certain Gram-positive and Gram-negative bacteria of bovine casein have also been demonstrated [19]. Casecidin 17 is well documented in the literature and several bioactivities have been suggested. Rojas-Ronquillo et al. found the angiotensin-converting enzyme inhibitory and antithrombotic properties of casecidin 17 with an inhibition efficiency ratio of 0.1%/peptide concentration (μg/ml) and 4.6%/peptide concentration (μg/ml), respectively [25]. Sandre et al. found that casecidin 17 has immunomodulatory activity in mice, probably by enhancing the antimicrobial activity of macrophages [26]. In terms of antibacterial properties, Birkemo et al. were the first to establish the inhibition effects of casecidin 17 of E. coli, indicated by in vitro MIC values of 0.4 mg/ml [27]. In this study, we found that the peptide extract mainly containing casecidin 17 and β-casein 207–224 had anti-H. pylori properties. Further studies will be focused on the anti-H. pylori activities of individual peptides.
As no antibacterial information on β-casein 207–224 can be found in various databases, we believe this study is the first to reveal its potential anti-H. pylori activities. β-casein 207–224 has only one more amino acid residue than does casecidin 17, and the rest of their sequences are in the same order. Hence, their structures and sequences are highly similar, which suggests that they may have similar biological functions. The reason for the small amount of information in the literature about β-casein 207–224 is that the hydrolysis of this peptide is not common in most experimental conditions. Here, the enzymolysis was performed in acidic environments, which could be an important factor for producing β-casein 207–224. As shown in Figures 1 and 2, the amount of β-casein 207–224 was much higher when the casein was enzymolyzed at pH 2.0, compared to at pH 3.0. Hence, β-casein 207–224 could be one of the major antibacterial peptides found in an empty human stomach, where the pH value is approximately 1.5 to 2.5. Furthermore, the MIC90 values at pH 2.0 are lower than those at pH 3.0, indicating that better anti-H. pylori activities were achieved in the pH 2.0 environment, where the concentration of β-casein 207–224 is higher (Figures 1 and 2).
Several studies have produced antibacterial peptide mixtures with casecidin 17 through different methods [25–27]. Our study is the first to use a simple in vitro pepsin enzymolysis method to successfully establish casecidin 17 in an extract. According to the supplier product information sheet (https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Product_Information_Sheet/c5890pis.pdf), the caseins used in this study were obtained from bovine milk and contain four main types of casein: α-s1 casein, α-s2 casein, β-casein and κ-casein. Pepsin is one of the major enzymes in the human gastrointestinal tract required for the digestion of ingested proteins, including milk. Hence, this in vitro study simulated the digestion process of bovine milk casein by pepsin, and we found that the major products of such a process are casecidin 17 and β-casein 207–224. A study showed that bovine milk contains about 32 g of protein per liter [28]. Casein has been considered to be the main constituent in milk and it makes up approximately 80% of the total protein in bovine milk [18].
The MIC90 values in this study were obtained from in vitro experiments; hence, many other in vivo factors that may affect the activity of the peptides were excluded. For example, there are other enzymes that may digest casein and produce other types of antibacterial peptides, which may not be casecidin 17 and β-casein 207–224. Another excluded factor is that the human gastrointestinal tract is highly complex and contains food and many other bacteria; hence, casecidin 17 and β-casein 207–224 may bind to other bacteria or other substances and have less direct contact with H. pylori. Nevertheless, this study provides the basis for further investigation on casecidin 17 and β-casein 207–224 for novel anti-H. pylori peptide design. Further optimization of antibacterial peptide extraction could assist in developing therapeutic agents to modulate the effect of antibiotics on H. pylori infections.