Cardiovascular diseases (CVDs) remain a leading cause of mortality and morbidity globally encompassing the ischemic heart disease, stroke, heart failure, peripheral arterial disease (PAD), and other heart and vascular conditions. With the growth and aging of populations, the incidence of CVDs rose sharply, which results in high mortality and readmission rates contributing significantly to healthcare burdens [1, 2]. Therefore, it is vital to find early biomarkers of CVDs. One research study proposes that serum enzymes such as alanine aminotransferase (ALT), aspartate aminotransferase (AST), and γ-glutamyl transferase (GGT) are potential predictors of CVDs [3] while another study has conflicting findings revealing no significant association between certain serum enzyme levels and CVDs [4, 5]. These discrepancies may stem from limited sample sizes, unaccounted confounding factors, or unclear causal relationships between exposures and outcomes. To obtain more accurate results, we first analyzed a large statistical sample through the National Health and Nutrition Examination Survey (NHANES) database, and then proceeded with the help of Mendelian Randomization (MR) to eliminate potential confounders that could affect the results. Our study evaluated the causal relationship between lactate dehydrogenase (LDH), ALT, AST, alkaline phosphatase (ALP), creatine phosphokinase (CK), GGT and CVDs, providing robust insights into the role of these serum enzymes in CVD pathogenesis.
Methods
This study was conducted in two distinct parts. The first part utilized data from the NHANES from 1999-2018 to examine the effects of multiple factors on CVDs. NHANES is a research program that evaluates the health and nutritional status of adults and children in the United States. Since 1999, NHANES has conducted an ongoing survey every 2 years. Each year, the research team conducts household interviews and physical examinations of approximately 5,000 nationally representative United States citizens. For this study, inclusion criteria comprised participants aged 20 years or older with complete CVD records and serum enzyme data. Individuals with missing data for key study variables or those who were pregnant were excluded. Indicators of serum enzymes in the NHANES database are in the Standard Biochemistry Profile for Laboratory Data and include ALT, AST, CK, GGT, ALP and LDH.
The second part was a two-sample MR analysis using summary statistics from the Genome-Wide Association Study (GWAS) to reveal the causal relationship between serum enzyme levels and CVDs. MR analysis views genetic variants as instrumental variables (IVs), aiming at evaluating the causality between exposure and outcome. Data for this analysis were derived from publicly available GWAS summary statistics. CVD data were sourced from the OPENWAGS of the Finnish GenBank R10, while serum enzyme data were obtained from the OPENGWAS database. We screened and processed the exposure GWAS data with the R language and selected single nucleotide polymorphism (SNP) that reached the threshold of genetic significance (p < 5 × 10–8) as IVs. SNP data for serum enzyme levels and CVDs were integrated and harmonized to ensure consistency of effector alleles. To ensure independence among SNPs, we applied the Clump method (parameter settings: kb = 500, r2 = 0.01) to eliminate linkage disequilibrium. In the case of chain imbalance, the SNP with the smallest p-value was retained as a representative. For missing SNPs in the outcome GWAS data, proxy SNPs were identified based on the criteria of r2 < 0.8 and MAF < 0.3 to ensure strong correlation with the target SNPs. In order to ensure reliability and accuracy of the analysis results, sensitivity analyses were performed by leave-one-out analysis and Cochran’s Q test. The leave-one-out analysis was used to check whether any individual variable had a significant influence on the overall results by excluding individual IVs one at a time. Cochran’s Q test was utilized to detect heterogeneity in the results. Additionally, to obtain more reliable conclusions, multivariate-adjusted forward MR analyses of smoking, drinking, and hypertension were performed to eliminate the effects of these confounding factors on causality.
Results
In the first study, a total of 37,411 individuals were included in the analysis during the NHANES data cycle, with a weighted analytic sample size representing approximately 160 million U.S. non-institutionalized participants. Among the included participants, 3,596 (9.612%) were diagnosed with CVDs. The association between six serum enzyme levels and CVDs was assessed with multivariate weighted logistic regression models as shown in Table I. The results showed that only CK (OR = 0.80, 95% CI = 0.69–0.93, p = 0.003) and LDH (OR = 2.07, 95% CI = 1.37–3.14, p < 0.001) were significantly associated with CVDs after adjusting for a wide range of confounders (age, sex, race, education, smoking, hypertension, diabetes).
Table I
Multi-factorial logistic regression analysis of the relationship between serum enzyme levels and CVDs
In the second part, in inverse variance-weighted (IVW) analysis, forward MR corrected for drinking, smoking, and hypertension, changes in ALT levels were associated with coronary atherosclerosis (OR = 0.854, 95% CI = 0.731–0.999, p = 0.048) and other peripheral vascular diseases (OR = 1.287, 95% CI = 1.024–1.617, p = 0.030), and ALP levels were significantly associated with atherosclerosis, excluding cerebral, coronary and PAD (OR = 0.856, 95% CI = 0.778–0.942, p = 0.002) (Figures 1 A–F). In the reverse MR, no significant causal relationship was observed between CVDs, ALP and ALT. These findings effectively rule out the possibility of reverse causality and further support the causal effect of serum enzyme levels on CVDs, as identified in the forward MR analysis. To evaluate the robustness and reliability of the results, multinomial evaluations and sensitivity analyses were performed. MR-Egger regression indicated that multinomial factors had a small influence on the causal estimates, with p-values of the intercept terms greater than 0.05. The weighted median analyses further corroborated the primary findings. Leave-one-out analyses and Cochran’s Q tests showed that the main analyses remained consistent after excluding any individual IVs, and no significant heterogeneity was found. In summary, we conclude that CK and LDH are statistically associated with CVDs, while ALP and ALT have a strong causal relationship with CVDs.
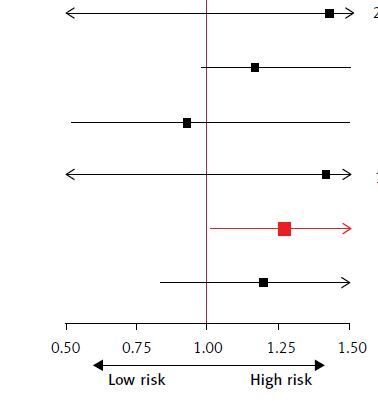
Figure 1
The forest plot presents the results of the multivariable MR IVW analysis. We investigated the causal relationship between six CVDs and serum enzymes using an inverse variance weighting/weighted method. The six CVDs represent A – Stroke. B – Atherosclerosis, excluding cerebral, coronary and PAD. C – Cerebral atherosclerosis D – Coronary atherosclerosis. E – Other peripheral vascular diseases. F – Embolic stroke. Statistically significant results are highlighted in red and error bands indicate 95% confidence intervals


Discussion
This study provides robust evidence linking serum enzyme levels to CVDs through a combination of large-scale observational data and MR analyses. By leveraging data from the NHANES and GWAS, we addressed limitations of prior research, including the small sample sizes and confounding variables, to clarify the causal roles of these serum enzymes in CVD pathogenesis.
Our findings highlight that compared to CK and LDH, which are statistically associated with CVDs, ALP and ALT also exhibit strong causal relationships with CVDs. These results are consistent with previous observational studies that demonstrate associations between serum enzyme levels and CVD risk, but lacked the rigorous methodology required to establish causality [5–7]. Wannamethee et al. found that ALP was related to coronary heart disease (CHD) risk in elderly men [6]. A systematic review conducted by Kunutsor et al. revealed a potential association between ALT levels and coronary heart disease (CHD) [5]. Additionally, studies investigating the relationship between CK levels and CVD risk have demonstrated that elevated CK levels are significantly associated with an increased risk of CVDs [8]. Similarly, an observational study by Zhu et al. reported that higher LDH levels are linked to a greater prevalence of atherosclerosis and CVDs [7]. Importantly, our forward MR analyses effectively ruled out reverse causality as no significant causal relationship was detected in the reverse MR analyses for ALP and ALT. This finding enhances the robustness of our results and underscores the potential role of these enzymes as biomarkers or mediators in the pathogenesis of CVDs.
Observational studies from the NHANES database show different outcomes from MR studies. This suggests that genetic variants may affect serum enzyme levels, and changes in these enzyme levels may influence the development of CVDs through complex gene-environment interactions. The link between serum enzymes and the presence of CVDs is very complicated; ALP acts through certain molecular mechanisms that directly contribute to vascular pathological processes [9–11]; GGT is present in atherosclerotic plaques, where it is associated with lipid peroxidation and atherosclerosis development [12, 13]; ALT and LDH may be secondary effects of inflammatory responses [14, 15]; CK is a specific biomarker of myocardial injury and is also affected by both vascular hemodynamics and energy metabolism [16], and AST may reflect myocardial injury but its cardiovascular relevance in the context of non-myocardial injury is not clear. Specifically, ALP may inhibit atherogenesis by improving intestinal barrier function and attenuating intestinal lipid absorption [9], meanwhile ALP also regulates pyrophosphate metabolism and activates intracellular signaling pathways such as bone morphogenetic proteins [10]. Hernández-Mosqueira et al. also found that ALP may upregulate FABP4, lipocalin gene expression and downregulate leptin expression [11]. When cells are injured, LDH release triggers inflammatory signaling that activates NLRP3 inflammatory vesicles, induces Caspase-1-dependent cellular pyroptosis, and disrupts endothelial barrier function [15]. Brewster et al. noted that CK is thought to be associated with blood pressure by affecting cardiovascular hemodynamics through enhancing systemic vascular resistance, stroke volume, and cardiac contractility [17]. ALT was significantly associated with coronary atherosclerosis and peripheral vascular diseases, while ALP was linked to atherosclerosis excluding cerebral and coronary conditions. These findings are particularly intriguing, given the dual roles of these enzymes in metabolic and inflammatory pathways. ALT, traditionally considered a marker of liver function, may reflect systemic metabolic disturbances that contribute to vascular dysfunction [18]. Similarly, elevated ALP levels, often associated with bone and liver disorders, have been increasingly recognized as an independent risk factor for vascular calcification and atherosclerosis [19].
Despite these strengths, several limitations should be acknowledged. First, although we minimized the effects of confounders and reverse causality through MR methods, limitations of data still remain. The association between genetic variation and phenotype may be affected by gene-environment interactions, which are difficult to fully account for in the analysis. Although we adjusted for multiple confounders, there may still be unrecognized or unmeasured confounders. Besides, the sample in this research was primarily from the United States population, and the results may not be fully applicable to populations of other races or regions.
In conclusion, changes in serum enzyme levels may reflect altered metabolic status of the body, these findings not only enhance our understanding of the pathophysiological mechanisms underlying CVDs but also offer potential avenues for early risk stratification and intervention. In addition, biochemical and molecular biology techniques are used to study the specific mechanisms of serum enzymes in the pathogenesis and development of cardiovascular disease and to analyze the role of these enzymes in gene expression and regulation.