Introduction
Emerging studies have demonstrated that the changes of protein post-translational modification in oncogenes (such as VEGFR1 and MAP3K2) and tumor suppressors (such as p53 and RB1) [1–5] play an important role in human tumorigenesis. The protein arginine methyltransferase (PRMT) family plays important roles in regulating the post-translational modification of multiple proteins, including histone proteins and non-histone proteins [6]. Emerging studies have indicated that PRMT family members were dysregulated and involved in regulating the progression of human cancers, such as breast cancer, and lung cancer. Among these PRMT family members, the important roles of PRMT1, PRMT2, PRMT5 and PRMT7 have been demonstrated in cancers [7].
Prostate cancer (PCa) is the most common male genitourinary malignancy in the world [8]. However, the biological mechanisms regulating PCa progression have remained elusive. Androgen receptor (AR) is the most important regulator involved in modulating PCa proliferation, invasion and glycolysis [9, 10]. Speckle-type POZ (SPOP) is one of the most frequently mutated genes in PCa, whose mutations were associated with drug resistance, lipid accumulation and genomic stability [11–13]. Recent studies showed PRMT family members regulated the tumorigenesis, progression and development of PCa through affecting multiple signaling pathways [14, 15]. For example, PRMT6 played an oncogenic role of in PCa [16]. PRMT10 is required for androgen-dependent proliferation of PCa LNCaP cells [17]. Therefore, to establish effective indicators of prognosis and therapeutic strategies against this disease, we need a clearer realization of the molecular pathology of PCa.
PRMT5 is a main member of the type II PRMT family [18]. In human cells, PRMT5 commonly interacts with MEP50 at the catalytic site and produces monomethylarginine [19]. Recent studies have indicated that PRMT5 plays a regulatory role in promoting cancer progression [20]. PRMT5 was observed to be overexpressed in multiple cancers, such as melanoma, lung cancer, ovarian cancer, breast cancer and prostate cancer [21]. More PRMT5 was expressed in PCa tissues, which is positively correlated with AR expression. PRMT5 promotes PCa cell growth by epigenetic activation of AR transcription [22]. Inhibition of PRMT5 down-regulates AR expression and inhibits the growth of multiple AR-positive PCa cells [23]. A previous study showed that ERG recruited PRMT5 to AR and methylate AR Arginine 761, which suppressed the recruitment and transcription of AR target genes [24]. However, the effects of PRMT5 on PCa metastasis and the up-stream regulators of PRMT5 in PCa remained to be further investigated.
In the present study, the expression of PRMT5 in PCa was analyzed by using public datasets and the molecular functions of PRMT5 in PCa were investigated using loss-of function assays. The aims of the study were to investigate whether PRMT5 could be a novel diagnostic marker and used as a therapeutic target in PCa, to explore the possible molecular mechanism, and to understand the clinical importance of PRMT5 in prostate carcinogenesis.
Material and methods
Cell culture
LNCaP, 22RV1, DU145, and PC-3 were purchased from the American Type Culture Collection (Manassas, VA) and confirmed by short tandem repeat (STR) analysis or mycoplasma detection, DNA fingerprinting, isozyme detection, and cell vitality detection. These cells were cultured in RPMI 1640 medium (with 10% fetal bovine serum, Biochrom) at 37°C and in a 5% CO2 humidified incubator for further study.
RNA interference
The siRNAs against the PRMT5 (siPRMT5-1, siPRMT5-2) and negative control (siNC) were synthesized by GenePharma (Shanghai, China). The PCa cells were seeded and transfected in 12-well plates using Lipofectamine 3000 (Life Technologies), in accordance with the manufacturer’s instructions. The following siRNAs were used: siPRMT5-1: 5′- CAACAGAGAUCCUAUGAUU -3′; siPRMT5-2: 5′- AAGAGGGAGUUCAUUCAGGAA -3′; and a scrambled siRNA control: 5′- AAGUGAUAGGAAGUCAGUACG -3′.
Plasmids and cell transfection
Human cDNA of PRMT5 was obtained from Jiahuai Han (Xiamen University) and cloned into plasmid pcDNA3.1(+) (Invitrogen). The PCa cells were seeded and transfected in 6-well plates with pcDNA3.1(+) or pcDNA3.1(+)-PRMT5 plasmid (800 ng/well) using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s instructions.
Real-time quantitative PCR
Beyozol (Beyotime, China) was used to extract total RNA from PCa cells. BeyoRT First Strand cDNA Synthesis Kit (Beyotime, China) was used to perform the reverse transcription according to the manufacturer’s instructions. Real-time quantitative PCR (RT-qPCR) was conducted using ABI Prism 7900HT (Applied Biosystems, Foster City, CA). Primers for PRMT5 were: forward, 5′- TGAATTGTCGCCTGAGTGC-3′ and reverse, 5′- GGGATGCTCACACCATCAT -3′. Primers for β-actin were: forward, 5′- GAGCTACGAGCTGCCTGACG -3′ and reverse, 5′- CCTAGAAGCATTTGCGGTGG -3′. Primers were synthesized by GenePharma (Shanghai, China). β-actin was selected as a reference. The 2–ΔΔCt method was used to calculate the relative mRNA expression. Each sample was run in triplicate [25].
Western blot analysis
Cells were harvested 48 h post transfection, using RIPA lysis buffer RIPA (strong). BCA Protein Assay Kit (Beyotime, China) was used to detect the content of proteins according to the manufacturer’s instructions. 12% (w/v) polyacrylamide gel SDS-PAGE was used to separate proteins. β-actin was selected as a reference. PRMT5 and ACTB were detected by monoclonal antibodies for PRMT5 and ACTB (1 : 1,000; Cell Signaling Technology, Beverly, MA, USA).
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) was used to measure cell proliferation according to the manufacturer’s instructions. After transfection, we measured the absorbance at 450 nm using a Microplate Reader at 0, 24, 48, 72, 96, and 120 h. The absorbance at 630 nm was selected as a reference. Each sample was run in triplicate.
Cell cycle and cell apoptosis assay
Cell Cycle and Apoptosis Analysis Kit (Beyotime, China) was used to perform the cell cycle assay according to the manufacturer’s instructions. The cells were harvested 48 h post transfection. 1 ml of pre-cool 70% ethanol was added and mixed by gently pipetting, then placed at 4°C for 12 h. Then the cells were collected by centrifugation at 1000 g, 3 min. The prepared Cell Cycle and Apoptosis Analysis Kit was added to each tube, and the cell pellet was slowly and fully resuspended, and left at 37°C for 30 min in the dark. The FACStar flow cytometer (Becton-Dickinson, San Jose, CA) was used to measure the cell cycle.
Annexin V-FITC Apoptosis Detection Kit (Beyotime, China) was used to perform the cell apoptosis assay according to the manufacturer’s instructions. The cells were harvested and gently resuspended by adding 195 μl of Annexin V-FITC binding solution. Then 5 μl of Annexin V-FITC and 10 μl of propidium iodide staining solution were added to the mixture and it was mixed gently. Then the mixture was incubated at room temperature for 15 min in the dark, then placed in an ice bath. A FACStar flow cytometer was used to measure the cell apoptosis.
Cell migration and invasion assay
Transwell plates (8-μm pore size, 6.5 mm diameter; Corning Life Sciences, Lowell, MA) with or without Matrigel (BD) were used to conduct cell migration and invasion assays according to the manufacturer’s protocol. For migration, 100 μl of medium containing 1.5 × 104 cells (1% FBS) was added to the upper chamber. 700 μl of medium (10% FBS) was added to the lower chamber.
ceRNA network construction
The lncRNA-miRNA interactions and miRNA-mRNA interactions were predicted using StarBase V2.0 (http://starbase.sysu.edu.cn/starbase2/). The ceRNA regulatory network was established based on lncRNA-miRNA-mRNA axes by combining lncRNA-miRNA interactions with miRNA-target gene interactions. Cytoscape v3.6.0 was used to visualize the ceRNA network.
Reporter constructs and luciferase assay
The Dual-Luciferase Reporter Assay System (Promega, USA) was used to perform the Luciferase assay. The predicted binding sites for miR-150 in ZFAS1 and 3’-UTR of PRMT5 were inserted into psi-CHECK2 Dual-Luciferase miRNA Target Expression Vector (Promega, USA) within the XhoI/NotI sites. Sequence integrity was confirmed by sequencing.
Statistical analysis
All data are shown as the mean ± standard deviation (SD). The SPSS software package, version 15.0 (SPSS Inc., Chicago, IL) was used to perform statistical analysis. Student’s t test was used to determine significant differences between two groups. In order to compare the significant differences among multiple groups (group ≥ 3), one-way ANOVA followed by Tukey’s test was used. Kaplan-Meier curves with log ranking tests were conducted to assess the correlation between PRMT5 or ZFAS1 and survival time in patients with PCa. A value of p < 0.05 was selected as significant.
Results
PRMT5 was overexpressed and correlated with poor prognosis of PCa
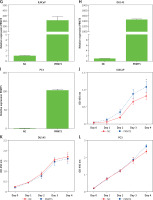
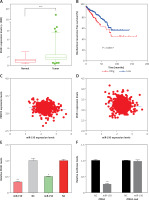
In order to further investigate the prognostic value of PRMT5 in PCa, we analyzed several public datasets, including TCGA and GEPIA. The results showed that the expression level of PRMT5 in PCa samples were significantly higher than that in normal prostate tissues by analyzing these datasets (Figures 1 A and B). Moreover, GSE21032 dataset analysis revealed that PRMT5 was up-regulated in metastasized PCa samples compared to primary PCa samples (GS < 7 PCa) and normal prostate tissues (Figure 1 C), suggesting that PRMT5 may be involved in regulating PCa metastasis.
Figure 1
PRMT5 was overexpressed and correlated with poor prognosis of PCa. A – TCGA and B – GEPIA datasets revealed that the expression level of PRMT5 in PCa samples was significantly higher than that in normal prostate tissues. C – GSE21032 dataset analysis revealed that PRMT5 was up-regulated in metastasized PCa samples compared to primary PCa samples (GS < 7 PCa) and normal prostate tissue. D – The BFS (p < 0.01) was higher in PRMT5-low patients compared to PRMT5-high patients. E – The OS (p < 0.05) rate was higher in PRMT5-low patients compared to PRMT5-high patients
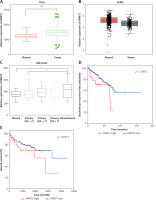
Furthermore, the association of PRMT5 expression with BCR-free survival (BFS) and overall survival (OS) in PCa patients was assessed by the Kaplan-Meier curve method. The cutoff point which divides all cases into PRMT5 high and PRMT5 low groups was the median PRMT5 mRNA expression in all PCa tissues. As shown in Figures 1 D and E, compared to PRMT5-high patients, the BFS (p < 0.01) and OS (p < 0.05) rates were higher in PRMT5-low patients in both datasets (p < 0.05, in both datasets), indicating that PRMT5 could be a potential biomarker for the prognosis of PCa.
Bioinformatics analysis of PRMT5 in PCa
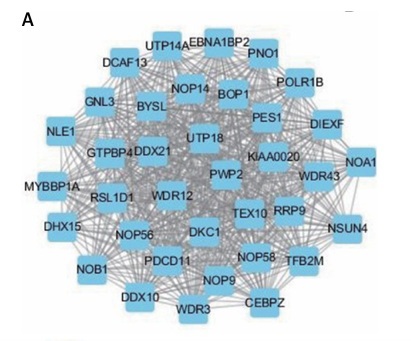
In order to explore the potential functions PRMT5 in PCa, the present study conducted co-expression analysis and protein-protein interaction (PPI) network analysis to identify the downstream targets of PRMT5. The top key up-regulated PPI (Figure 2 A) and down-regulated PPI networks (Figure 2 C) are shown in Figure 2. Bioinformatics analysis revealed that PRMT5 was involved in regulating multiple biological processes and pathways, such as cell junction assembly, angiogenesis, cell−cell adhesion, cell proliferation, and spliceosome (Figures 2 B and D). These analyses were consistent with previous studies indicating that PRMT5 could promote PCa proliferation.
Figure 2
Bioinformatics analysis of PRMT5 in PCa. A – The top key up-regulated protein-protein interaction (PPI) networks of the downstream targets of PRMT5. B – The top key down-regulated PPI networks of the downstream targets of PRMT5. C – The biological processes and pathways involved in the top key up-regulated PPI networks D – The biological processes and pathways involved in the top key down-regulated PPI networks

PRMT5 promotes proliferation of androgen-dependent PCa, not androgen-independent PCa
The mRNA levels of PRMT5 were evaluated in the normal prostate WPMY-1 cells and human PCa cell lines, including LNCaP, DU145, and PC-3 by qRT-PCR analyses, respectively (Supplementary Figure S1 C). Our results showed that mRNA levels of PRMT5 were significantly induced in PCa LNCaP, 22RV1, DU145, and PC-3 compared to normal WPMY-1 cells. These results suggested that the overexpression of PRMT5 might be positively correlated with the progression of prostate cancer.
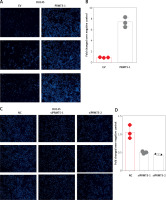
To characterize the function of PRMT5 in PCa, PRMT5 expression was knocked down at the mRNA and protein levels by PRMT5 siRNA. PRMT5-specific siRNA substantially decreased PRMT5 mRNA and protein levels in PCa cells compared with the siRNA control (NC) (Figures 3 A–C). Then, the present study detected the effect of PRMT5 on cell proliferation in PCa using the CCK-8 assay. The results showed that the proliferation rate of the androgen-dependent PCa LNCaP cell line was significantly reduced after transfection with siPRMT5-1 and siPRMT5-2 compared to cells treated with the siNC (p < 0.001, Figure 3 D). Interestingly, our results showed that knockdown of PRMT5 did not affect the cell proliferation of androgen-independent PCa DU145 and PC-3 cells (Figures 3 E and F).
Figure 3
PRMT5 promotes proliferation of androgen-dependent PCa, not androgen-independent PCa. A–C – PRMT5-specific siRNA-1,2 substantially decreased PRMT5 mRNA levels in PCa cells LNCaP, DU145 and PC3. D – Knockdown of PRMT5 significantly reduced the proliferation rate of androgen-dependent PCa LNCaP cell line compared to cells treated with siNC (p < 0.001). E, F – Knockdown of PRMT5 did not affect the cell proliferation of androgen-independent PCa DU145 and PC-3 cells G–I – Overexpression of PRMT5 significantly increased PRMT5 gene expression in PCa cells LNCaP, DU145 and PC3. J – Overexpression of PRMT5 remarkably enhanced proliferation of LNCaP cells (p < 0.05). K, L – Overexpression of PRMT5 did not significantly promote DU145 and PC3 proliferation
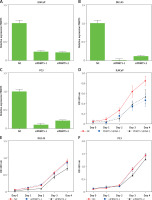
Subsequently, the present study validated the above finding through overexpressing PRMT5 in PCa cells. After the plasmid containing PRMT5 gene was transfected in PCa, a significant increase of PRMT5 gene expression was detected (Figures 3 G–I). Our results showed that overexpression of PRMT5 remarkably enhanced the proliferation of LNCaP cells (p < 0.05, Figure 3 J) and did not significantly promote DU145 and PC3 proliferation (Figure 3 K and L). Taken together, these results showed that PRMT5 promotes proliferation of androgen-dependent PCa, not androgen-independent PCa.
PRMT5 promotes the cell cycle progression of androgen-dependent PCa, not androgen-independent PCa
Next, the effect of PRMT5 on the cell cycle progression of LNCaP and DU145 cells was assessed by flow cytometry. As presented in Figure 4, the results showed that knockdown of PRMT5 in LNCaP cells significantly increased the percentage of G1 phase cells and reduced the percentage of S and G2/M phase cells compared to the NC group (p < 0.001; Figure 4 A). However, our results showed that silencing of PRMT5 in DU145 cells did not change the percentage of G1 phase cells and S phage cells compared to the NC group (Figure 4 B). Furthermore, the present study found that PRMT5 overexpression significantly promoted the cell cycle by reducing G1 phase and inducing S phase cells in LNCaP cells, not in DU145 cells.
Figure 4
PRMT5 promotes cell cycle progression and cell apoptosis of androgen-dependent PCa, not androgen-independent PCa. A – Knockdown of PRMT5 in LNCaP cells significantly increased the percentage of G1 phase cells and reduced the percentage of S and G2/M phase cells compared to NC group (p < 0.001). B – Knockdown of PRMT5 in DU145 cells did not change the percentage of G1 phase cells and S phage cells compared to NC group. C – Knockdown of PRMT5 significantly increased the early and late apoptotic cell fractions by 18% and 37.5% in LNCaP cells, respectively D – Knockdown of PRMT5 had no effect on the androgen-independent DU145 apoptosis

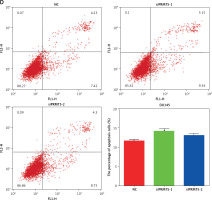
PRMT5 suppressed cell apoptosis of androgen-dependent PCa
Cell apoptosis plays crucial roles in regulating cancer proliferation. The present study conducted Annexin V-FITC/PI assay to detect the effect of PRMT5 on cell apoptosis in PCa. As shown in Figure 4 C, knockdown of PRMT5 using siPRMT5-1 and siPRMT5-2 significantly increased the early and late apoptotic cell fractions by 18% and 37.5% in LNCaP cells, respectively. However, knockdown of PRMT5 had no effect on the androgen-independent DU145 apoptosis (Figure 4 D).
PRMT5 promotes cell migration and invasion of androgen-independent PCa
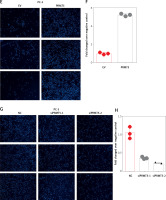
Considering that PRMT5 did not affect the proliferation of androgen-independent PCa, the present study hypothesized that PRMT5 might be involved in promoting androgen-independent PCa metastasis. First, the present study performed transwell assay to detect the effect of PRMT5 overexpression on PC-3 and DU145 cell migration. Our results showed that overexpression of PRMT5 remarkably enhanced PC-3 and DU145 cell migration ability (Figures 5 A, B, E, F). Of note, the present study found that knockdown PRMT5 significantly decreased PC-3 and DU145 cell migration ability compared to control groups in PC-3 and DU145 cells (Figures 5 C, D, G, H).
Figure 5
PRMT5 promotes cell migration of androgen-independent PCa. Overexpression of PRMT5 remarkably enhanced PC-3 (A, B) and DU145 (E, F) cell migration ability. Knockdown of PRMT5 significantly decreased PC-3 (C, D) and DU145 (G, H) cell migration ability compared to control groups
Furthermore, the present study detected the effect of PRMT5 on cell invasion in PCa by estimating the penetration of cells through Matrigel in a transwell chamber. As illustrated in Figure 6, the present study found that knockdown of PRMT5 significantly inhibited cell invasion in DU145 and PC-3 (Figure 6 A) and PC-3 cells (Figure 6 C). The numbers of invading cells were decreased in PC-3 and DU145 cells transfected with siPRMT5, compared with the negative control group (p < 0.001, Figures 6 B and D).
Figure 6
PRMT5 promotes cell invasion of androgen-independent PCa. Knockdown of PRMT5 significantly inhibited cell invasion in DU145 (A, B) and PC-3 cells (C, D)
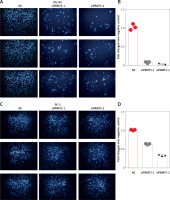
These results indicated that PRMT5 significantly increased invasion, and migration of androgen-independent PCa cells.
Identification of the potential competing endogenous RNA networks involved in PRMT5 regulation in PCa
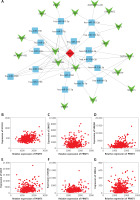
The upstream regulators of PRMT5 in PCa remained largely unknown. Long non-coding RNAs had been demonstrated as a class of key important regulators of PCa progression. The present study for the first time constructed the potential competing endogenous RNA networks involved in PRMT5 regulation in PCa. The present study first conducted a co-expression network based on the correlation analysis between the PRMT5 and differently expressed lncRNAs. The present study selected the PRMT5-lncRNA pair with the Pearson correlation coefficient > 0.4 for further study. Then, the present study predicted potential miRNAs targeting the PRMT5-lncRNA pair using StarBase, TargetScan, and miRcode. Then, the competing endogenous RNA networks were presented using Cytoscape software.
As shown in Figure 7 A, a total of 17 lncRNAs (including GAS5, SNHG3, SNHG7, ZFAS1) and 26 miRNAs were included in this network. The most significantly co-expressing lncRNAS of PRMT5 in PCa included HCG18, SNHG7, SNHG8, GAS5, ZFAS1, and SNHG3 (Figures 7 B-G).
Figure 7
The potential competing endogenous RNA networks involved in PRMT5 regulation in PCa. A – A total of 17 lncRNAs (including GAS5, SNHG3, SNHG7, ZFAS1) and 26 miRNAs were included in this network. B–G – The most significantly co-expressing lncRNAS of PRMT5 in PCa included HCG18, SNHG7, SNHG8, GAS5, ZFAS1, and SNHG3
The present study focused on the ZFAS1/miR-150-5p axis in PCa. ZFAS1 and miR-150-5p had been reported to be dysregulated and involved in the regulation of human cancer progression. However, their roles in PCa remained unclear. This study for the first time showed that ZFAS1 was significantly overexpressed in PCa and correlated with shorter disease-free survival time in patients with PCa (Figures 8 A and B). Moreover, the present study found that the expression levels of miR-150-5p were negatively correlated with both ZFAS1 and PRMT5 expression levels in PCa (Figures 8 C and D). These results suggested that the FAS1/miR-150-5p axis may participate in the regulation of PRMT5 expression in PCa.
Figure 8
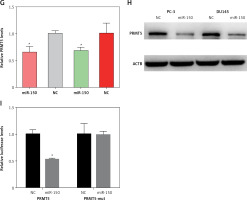
ZFAS1 acted as a ceRNA to up-regulate PRMT5 expression though miR-150-5p. A – ZFAS1 was significantly overexpressed in PCa. B – ZFAS1 was correlated with shorter disease-free survival time in patients with PCa. C – The expression level of miR-150-5p was negatively correlated with ZFAS1 expression levels in PCa. D – The expression level of miR-150-5p was negatively correlated with PRMT5 expression levels in PCa. E – Overexpression of miR-150-5p significantly reduced the RNA levels of ZFAS1 in PC-3 and DU145 cells. F – Overexpression of miR-150-5p significantly decreased the luciferase activity of ZFAS1 G – Overexpression of miR-150-5p significantly reduced the RNA levels of PRMT5 in PC-3 and DU145 cells. H – Overexpression of miR-150-5p significantly reduced the protein levels of PRMT5 in PC-3 and DU145 cells. I – Overexpression of miR-150-5p significantly decreased the luciferase activity of PRMT5
ZFAS1 acted as a ceRNA to up-regulate PRMT5 expression though miR-150-5p
Then, the present study detected the expression levels of ZFAS1 and PRMT5 after overexpressing miR-150-5p in PC-3 and DU145 cells. As presented in Figures 8 E and G, our results showed that the RNA levels of ZFAS1 and PRMT5 were significantly reduced after overexpressing miR-150-5p in PC-3 and DU145 cells using RT-PCR assay. Western blot assay also showed that the protein levels of PRMT5 were significantly decreased in PC-3 and DU145 cells transfected with miR-150-5p compared to normal samples (Figure 8 H).
Furthermore, the direct interaction between miR-150-5p and PRMT5 or ZFAS1 was detected using luciferase assays. The luciferase reporter assay revealed that luciferase activity was significantly repressed in the constructs of the ZFAS1 (Figure 8 F) and PRMT5 -3’UTR (Figure 8 I) when co-transfected with the corresponding miRNAs compared with NC, whereas the mutated 3’UTR did not show a significant response to miR-150-5p.
ZFAS1 promoted cell migration and invasion of PCa though PRMT5
As presented in Figures 9 A and B, the present study found that knockdown of ZFAS1 significantly inhibited migration of PC-3 5 cells by 75% compared with the NC group, respectively. Moreover, knockdown of ZFAS1 significantly inhibited invasion of PC-3 cells by 47% compared with the NC group (Figures 9 C and D), respectively.
Figure 9
ZFAS1 promoted cell migration and invasion of PCa though PRMT5. A, B – Knockdown of ZFAS1 significantly inhibited migration of PC-3 cells compared with NC group; overexpression of PRMT5 could rescue the migration. C, D – Knockdown of ZFAS1 significantly inhibited invasion of PC-3 cells compared with NC group; overexpression of PRMT5 could rescue the invasion
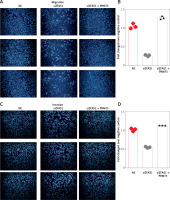
To test whether ZFAS1 promoted cell migration and invasion of PCa though PRMT5, the present study conducted rescue experiments. PC-3 cells co-transfected with siZFAS1 and PRMT5 significantly enhanced the PC-3 cell migration ability compared to the cells only transfected with siZFAS1 (Figures 9 A–D).
Discussion
PRMT5 was regarded as an oncogene in human cancers by regulating multiple cancer related regulators, such as TP53 and PDCD4 [26]. For example, the methylation of p53 by PRMT5 inhibits the expression of pro-apoptotic and anti-proliferative genes, thereby maintaining tumor cell self-renewal and proliferation. The methylation of E2F1 mediated by PRMT5 suppressed its activity to inhibit cancer proliferation [27]. PRMT5 has been observed to be overexpressed in various human cancers, such as melanoma, lung cancer, ovarian cancer, breast cancer and prostate cancer [21]. Of note, recent studies indicated that PRMT5 plays crucial roles in PCa tumorigenesis and development though affecting AR. AR is the most important regulator in PCa progression. Previous studies showed that PRMT5 promotes PCa cell growth by epigenetic activation of AR transcription. Inhibition of PRMT5 down-regulates AR expression and inhibits the growth of multiple AR-positive PCa cells [22, 24]. However, the effects of PRMT5 on PCa metastasis remained unclear. The present study conducted loss/gain of function assay to evaluate the roles of PRMT5 in PCa. Our results showed that PRMT5 significantly promoted androgen-dependent PCa proliferation and cell cycle progression and suppressed cell apoptosis. However, PRMT5 did not affect androgen-independent PCa proliferation. These results were consistent with previous reports and AR may play a key role in the regulation of PRMT5 functions in androgen-dependent PCa. Very interestingly, this study showed that PRMT5 could significantly induce androgen-independent PCa metastasis. Knockdown of PRMT5 suppressed, whereas overexpression of PRMT5 induced, cell migration and invasion in androgen-independent DU145 and PC-3 cells.
The up-stream regulators of PRMT5 remained largely unclear. A previous study showed that nuclear factor Y (NF-Y) transcriptional activation of PRMT5 is required for cell growth and is regulated by PKC /c-Fos signaling in prostate cancer cells [21]. Several miRNAs, such as miR-92b and miR-96, were also reported to be involved in the post-transcriptional regulation of PRMT5 in human cancers [28]. The present study for the first time showed that long non-coding RNAs were also involved in promoting PRMT5 expression in PCa though ceRNA mechanisms. LncRNAs are a novel class of RNA transcripts longer than 200 nucleotides. LncRNAs were found to be dysregulated in multiple cancers and regulated cancer related processes though binding to miRNAs, protein and DNAs. For example, lncRNA HULC promote PCa progression [29], lncRNA PVT1 promote multiple myeloma progression by inhibiting miR-203a expression [30], and lncRNA MIAT promoted epithelial ovarian cell proliferation and inhibited cell apoptosis by inhibiting miR-330-5p expression [31]. The most well-known mechanism of lncRNAs regulating targets is the ceRNA mechanism [32]. Also, ceRNAs show importance in regulating cancer progression [33]. For example, SNHG20 promotes PCa cell proliferation and migration via acting as a ceRNA to upregulate DDX17 [34]. The present study conducted bioinformatics analysis and constructed a ceRNA network involved in regulating PRMT5 expression in PCa. The further validation showed that ZFAS1 could up-regulate PRMT5 expression though sponging miR-150-5p.
LncRNA ZFAS1 had been reported to be up-regulated in several types of cancers, including hepatocellular carcinoma, colorectal cancer, gastric cancer, and glioma [35]. For example, ZFAS1 promotes tumorigenesis through regulation of miR-150-5p/RAB9A in melanoma [36]. Knockdown of ZFAS1 suppresses the progression of acute myeloid leukemia by regulating microRNA-150/Sp1 and microRNA-150/Myb pathways [37]. SP1-induced ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis [38]. In ovarian cancer, ZFAS1 was reported to promote ovarian cancer cell malignancy by interacting with miR-150-5p [39]. These reports were consistent with our finding that ZFAS1 could interact with miR-150-5p in PCa. Of note, this study for the first time demonstrated that ZFAS1 acted as a metastasis promoter in PCa. Knockdown of ZFAS1 could significantly reduce PCa migration and invasion. Moreover, our study showed that ZFAS1 promoted cell migration and invasion of PCa though PRMT5. Interestingly, our study analyzed the expression levels of ZFAS1, miR-150-5p and PRMT5 in PCa using a TCGA dataset. Our results showed that ZFAS1 and PRMT5 were overexpressed and miR-150-5p was down-regulated in PCa samples. Higher expression of ZFAS1 and PRMT5 was correlated with shorter disease free survival time in PCa patients. Moreover, the present study found that the expression levels of miR-150-5p were negatively correlated with both ZFAS1 and PRMT5 expression levels in PCa. These results suggested that the ZFAS1/miR-150-5p axis may participate in the regulation of PRMT5 expression in PCa.
In conclusion, the present study showed that PRMT5 promoted androgen-dependent PCa proliferation and androgen-independent PCa migration and invasion. PRMT5 knockdown decreased and PRMT5 overexpression enhanced DU145 and PC3 migration and invasion. Moreover, our results showed that the ZFAS1/miR-150-5p axis regulated PRMT5 expression in PCa cells. Furthermore, the present study showed ZFAS1 and PRMT5 were overexpressed and miR-150-5p was down-regulated in PCa samples. Higher expression of ZFAS1 and PRMT5 was correlated with shorter disease free survival time in PCa patients. These results showed that PRMT5 may be a therapeutic target for PCa.