In 1997, the presence of cell-free fetal (cff-) DNA in maternal plasma was demonstrated, ushering in a new era of noninvasive prenatal testing (NIPT) [1]. Over the past two decades, this burgeoning revolutionary strategy has considerably improved the early detection of birth defects worldwide [2]. Currently, cff-mRNA is emerging as a promising biomarker for NIPT beyond cff-DNA, as it can not only be used to screen for genetic defects such as aneuploidy but also be theoretically considered to have far more potential than DNA for real-time monitoring of fetal physiopathology [3]. However, cff-mRNA is susceptible to degradation and lacks stability and integrity as a marker, which greatly limits the feasibility of its clinical application [4]. Therefore, searching for a novel type of stable cff-RNA in the maternal circulation is particularly important for NIPT to break through the current dilemma. Here, for the first time, we report the presence of a stable RNA derived from fetal circulation, cff-circRNA, in maternal circulation, which might have implications for the NIPT strategy to monitor fetal biological alterations.
Methods
Sample collection and processing
The pregnant women who came to the First Hospital of Chongqing Medical University for a delivery hospitalization were recruited. All of them had singleton fetuses and were without serious pregnancy complications such as preeclampsia and fetal distress. With informed consent, their peripheral blood was collected in the third trimester (gestational week: 34.6 ±4.0) and after delivery using EDTA anticoagulant tubes. Then, the blood samples were centrifuged twice at 3000 rpm at 4°C to obtain plasma and stored at –80°C until testing [5]. Based on the pregnancy outcome, we finally selected samples from seven pregnant women carrying male fetuses for this study. The clinical information of these patients is shown in Supplementary Table SI.
The study was approved by the Medical Research Ethics Committee of the First Affiliated Hospital of Chongqing Medical University.
CircRNA microarray analysis
Total RNAs were extracted from plasma samples and purified using Trizol reagent (Invitrogen, Gaithersburg, MD, United States) and the NucleoSpin R RNA clean-up kit (MACHEREY-NAGEL, Germany), respectively. Then, the linear RNAs were removed by RNase R (Epicentre, Illumina, Inc.). The remaining RNAs were sequentially processed as follows: 1) reverse transcribed to first strand cDNA; 2) second strand cDNA synthesized; 3) cRNA synthesized using T7 Enzyme Mix; 4) reverse transcribed to cDNA; 5) reacted with Random Primer; 6) labeled with dNTP (Cy3-dCTP, Cy5-dCTP) with fluorescent moieties. The DNA with fluorophore was hybridized with the circRNA microarray in the hybridization mixture. Finally, the microarray was scanned using an Agilent microarray scanner (G2565CA); data were extracted using Agilent Feature Extraction (version = 10.7) software and normalized using Agilent GeneSpring software. All the processes of microarray analysis were conducted by the Bioassay Laboratory of CapitalBio Corporation (Beijing, China). The detailed information of experimental procedures is listed in Supplementary material.
RNA-seq data collection and analysis
We searched public databases for fetal/newborn peripheral blood sequencing datasets. Inclusion criteria were as follows: 1) the blood sample was collected immediately after birth for sequencing; 2) the information on the sex of the sample donors was provided; and 3) the data were in *.fastq format. Ultimately, the RNA-seq raw data of peripheral blood from male and female newborns were obtained from the SRA (https://www.ncbi.nlm.nih.gov/sra) database (accession ID: SRP182878).
The fastq files were processed by the BWA-MEM algorithm to map the reads to the UCSC human reference genome (hg19). Then, the circRNAs were identified and quantified using the CIRI2 software, which works based on the recognition of the back-spliced junction reads. All the processes were used with the default parameters.
The count data of circRNA were normalized by the TMM algorithm of the R package “edgeR”.
Ethics
The studies involving human participants were reviewed and approved by the Medical Research Ethics Committee of The First Affiliated Hospital of Chongqing Medical University (ID: 2020-60) and were subject to the Declaration of Helsinki. The patients/participants provided their written informed consent to participate in this study.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism (version 8.0, San Diego, CA, United States). The relative expression levels of cff-circRNAs were represented by the total concentration of the circRNAs derived from the Y chromosome (Y-circRNA). The visualization of results was performed by GraphPad Prism and the R package “gglpot2”.
Results
The detection of RNA and DNA derived from the Y chromosome of pregnant women with male fetuses demonstrated the presence of cff-RNA and cff-DNA in the maternal circulation, which led to the discovery of NIPT [1, 6]. With this, we speculate that if circRNA derived from the Y chromosome (Y-circRNA) can be found in the maternal circulation, it may further promote the development of NIPT. To confirm its feasibility, we analyzed circRNA expression profiles in peripheral blood of male and female neonates using RNA-seq data. The results showed that Y-circRNA was detectable in all the male neonates (n = 19), while it was negative in all the female samples (n = 12) (Supplementary Table SII). Thus, the presence of cff-circRNA can be proved by the detection of Y-circRNA in the maternal circulation.
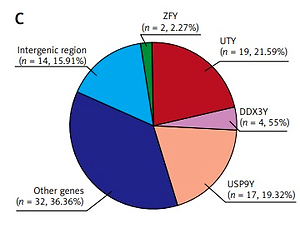
Based on this, we examined the cff-circRNA in the circulation of 7 pregnant women with a male fetus before delivery via a microarray targeting Y-circRNA. Of the total 198 Y-circRNAs to be measured, 88 were detected in at least 3 samples, and 27 of them were detected in all 7 samples (Figures 1 A, B). This indicates that cff-circRNA does exist in the maternal circulation, and most of these detected Y-circRNAs are derived from the genes UTY, USP9Y, DDX3Y and ZFY, which is consistent with our findings in the peripheral blood of male newborns mentioned above (Figure 1 C).
Figure 1
Characterization of Y-circRNA in maternal circulation. Y-circRNA expression profiles in (A) prenatal and (D) postpartum maternal plasma. B – Statistics of the counts of Y-circRNAs detected in prenatal maternal plasma samples. C – The percentage of Y-circRNA of different genetic origins in the prenatal maternal circulation (left panel) and in the neonatal circulation (right panel). E – The intersection of Y-circRNAs in prenatal and postnatal maternal plasma
Is the cff-circRNA in the maternal circulation sufficiently stable? In view of the instability of cff-mRNA in the circulation (the half-life is only 14 min), and the fact that it is quickly cleared after delivery [7], we performed the same test on the plasma of 7 pregnant women 24 h after delivery. The results showed that 66 of the 88 cff-circRNAs in prenatal plasma were still detectable in more than 3 postpartum plasma samples (Figures 1 D, E). This indicates that the existence of cff-circRNA (at least Y-circRNA) in the maternal circulation is stable, and may even last for a long time after delivery.
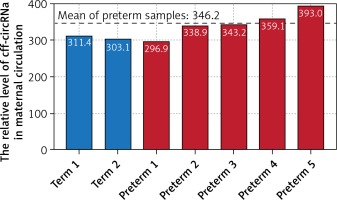
What is the significance of these circRNAs that are stably present in the maternal circulation? We preliminarily analyzed the association between cff-circRNA levels and clinical phenotypes to explore whether cff-circRNA may reflect underlying maternal-fetal biological alterations. We found that the relative expression levels of cff-circRNAs (represented by total concentrations of Y-circRNA) were higher in the prenatal plasma of pregnant women who eventually delivered prematurely than in that of full-term pregnancies (Figure 2). This suggested that more cff-circRNA may be transported from fetal to maternal circulation in the presence of pathological changes in the maternal-fetal system. That is, cff-circRNA might have potential as a biomarker for prenatal disease screening.
Discussion
In this study, we have demonstrated for the first time the presence of cff-circRNA in maternal plasma and found its potential association with diseases during pregnancy. This is a pioneering study and the results are preliminary, so large-scale clinical research is needed to validate and explore them in depth. A further limitation is related to the difficulties prevailing in current circRNA research. As a “young” RNA molecule, circRNA’s recognition algorithm and naming system are far less mature than those of traditional RNAs, and there is even some confusion. This poses a challenge to our research. Nevertheless, our findings have good clinical application and promotion value. Firstly, circRNA is an emerging RNA with a unique closed-loop structure distinct from traditional linear RNAs (mRNA, miRNA, lncRNA) and thus has the property of stable existence against nucleases [8, 9]. So our findings have the potential to help solve one of the biggest shortcomings of the current clinical application of cff-RNA. Secondly, compared with traditional linear RNA, circRNA has more significant tissue and spatiotemporal specificity; thus, in theory, cff-circRNA may reflect fetal pathological alterations more accurately [10, 11]. Thirdly, current studies on circRNA in maternal blood do not take into account the presence of cff-circRNA, meaning that cff-circRNA is also counted and analyzed as maternal circRNA. This might introduce potential errors into the study results. So, it is valuable to draw the attention of researchers to this issue via our findings.
In conclusion, we have identified a new type of fetal nucleic acid in the maternal circulation that may shed new light on the development of NIPT techniques and influence the achievement of real-time monitoring of fetal pathological alterations. It deserves to be explored by more systematic and extensive studies.