Coronavirus disease 2019 (COVID-19) can complicate other organs besides the lungs, including the brain, resulting in a series of neurological symptoms from headache to psychological disorder, cognitive impairment and even consciousness loss, especially in severely infected patients in the intensive care unit (ICU) [1]. A series of studies reported various neurological symptoms in COVID patients in the ICU, with an increasing rate since then [2–5]. However, the mechanisms of neuronal dysfunction in severe COVID-19 are not entirely known [2–4].
A recent study showed that 581 out of 4711 (12%) COVID patients had neurologic issues and patients with abnormal mentation and stroke had a greater risk of mortality [5]. However, the mechanisms of neuronal dysfunction in severe COVID-19 are not entirely known. Given the pathophysiology of inflammation storm and immunological deficit, we could infer that such a malfunction might be produced by an systemic inflammation together with increased permeability, causing blood-brain barrier (BBB) impairment. Accordingly, the invasion of cytokines and inflammatory factors into the brain is much easier, and direct viral entrance into the central nervous system (CNS) is less likely [6–8]. Detection of brain dysfunction, typically underdiagnosed in critically ill patients [9], might constitute a hurdle in the course of severe COVID-19.
Previously, we carried out gene set enrichment analysis (GSEA) in patients with traumatic brain injury (TBI) to detect a downregulated immune system, including Th1, Th2, and Th17 cell differentiation, and natural killer (NK) cytotoxicity in severe TBI [10]. This is consistent with the GO and KEGG results showing that the most enriched pathways were Th1 and Th2 cell differentiation, Th17 cell differentiation, hematopoietic cell lineage, and T cell receptor signaling pathway, which indicates that the immune deficiency is highly critical in brain insults. Accordingly, we obtained similar findings in severe COVID patients, in which the peripheral status demonstrates immune suppression and dominant inflammation. Therefore, we aimed to explore the association between COVID and ischemic stroke regarding the peripheral gene alterations.
Methods
Data processed
No human or animal experiments were involved in this study. We downloaded the datasets (GSE172114, GSE157103) from the NCBI website [11, 12]. GSE172114 has ninety-two blood samples including 46 COVID patients in the ICU and 23 non-ICU COVID patients. The GSE157103 dataset has 50 COVID patients in the ICU, and 50 non-ICU COVID patients. For the stroke datasets, we downloaded the datasets (GSE58294, GSE16561) from the NCBI website [13, 14]. GSE58294 (GPL570) has ninety-two blood samples including 69 ischemic stroke patients and 23 healthy controls. The GSE16561 dataset (GPL6883) has 39 ischemic stroke patients and 24 healthy controls. GSE22255 has 20 ischemic stroke patients and 20 healthy controls were used as the external validated datasets. We also included the GSE98895 dataset, which had 20 patients with metabolic syndrome and 20 healthy controls.
Single cell sequencing data obtained and processing
The single-cell data were downloaded from the NCBI website: GSE171555. The CellChat R package (1.4.0) was applied to explore the cell-cell interaction between the cell clusters. The R package monocle v2.22.0 was applied for pseudo-time analysis to get the genes from different pathological stages.
Results
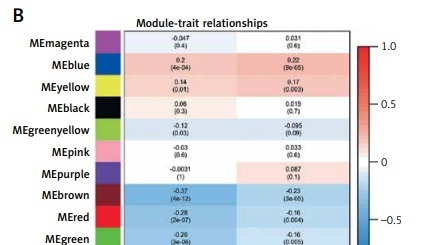
To investigate the differential genes in COVID and stroke, we crossed these genes and obtained 19 co-increased genes and 24 co-decreased genes in both COVID and stroke (Supplementary Figure S1). In addition, we combined these genes and applied WGCNA to link these genes with clinical characteristics (Supplementary Figure S2 A–C). We obtained 12 modules (including one grey module, Figure 1 A). According to the module-trait relation figure, we found that the MEblue module has a positive relationship with both severe COVID (sCOVID) (r = 0.2, p = 4e-4) and stroke (r = 0.22, p = 9e-5), and this indicated that these genes might be involved in both diseases. The correlation between gene significance and module genes was 0.25, p = 4.8e-15 and 0.29, p = 4e-09 in COVID and stroke, respectively (Supplementary Figures S2 D–H).
Figure 1
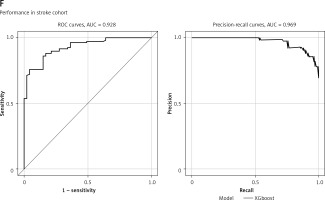
Shared hub genes in COVID and stroke. A – WGCNA analysis shows the modules related to both COVID and stroke. B – Module-trait relationships for the COVID-based WGCNA and stroke-based WGCNA. C – The importance plot of random forest shows the prediction of patients with severe COVID (red lines), patients without severe COVID (green lines) and the potential prediction error (black lines). D – The gene rank in random forest based on mean decrease of Gini value. E – ROC and PRC for sCOVID and stroke. F – ROC and PRC for sCOVID and stroke
To construct the diagnostic model, we applied the random forest method in the combing module genes in WGCNA and 43 DEGs with the “randomForest” package, and obtained the importance plot and the 30 gene ranks based on the mean decreased Gini value (Figures 1 B–D).
The predictive ability of each gene in the COVID dataset is listed in Supplementary Figure S3. The diagnostic model was also validated in the sCOVID testing model and stroke2 with AUC at 0.893 and 0.938, respectively, while the PRC value is 0.909 and 0.934, respectively (Supplementary Figures S2 I, J).
In addition, we found several shared pathways in these two diseases, dominated by the same genes. GRB10 is positively associated with hypoxia, complement, and glycolysis in both diseases, while CCR5 is negatively associated with these pathways in both COVID and stroke (Supplementary Figure S4).
To further clarify the role of stroke-related genes in sCOVID, we analyzed the sc-RNA seq data from GSE 171555. We first did the cell annotations in both groups (Supplementary Figure S5 A) and compared the complement and hypoxia extent in the PBMC between normal and severe COVID patients in different cell groups (Supplementary Figures S5 A–C). As both the complement and hypoxia levels were obviously enriched in monocytes, we further took them for re-clustering (Supplementary Figure S4 B), which showed that the sCOVID group had a higher level of both complement and hypoxia compared to the normal COVID group (Supplementary Figures S5 D–H).
As stroke-related genes in COVID have a critical role in immune infiltration assessed by CIBERSORT, we further explored the cell-cell interaction at a single cell level with the CellChat R package. We first found the close interaction between several cell types (Supplementary Figures S6 A–D). In addition, we observed the interaction between monocytes with high or low complement and other cell types in both outgoing and incoming signaling patterns (Supplementary Figure S6 E).
We further linked the risk gene expression with the complement and hypoxia score. We found that both GRB10 and CCR5 were mostly enriched in the monocytes (Supplementary Figure S7 A); and they were further closely associated with the complement and hypoxia score in monocytes (Supplementary Figures S7 B–F). We then applied pseudo-time analysis to identify the transition of monocyte sub-clusters, and we could see a clear stage change of monocytes from severe status to the non-severe status and relevant alteration of MCEMP1 during the transition (Supplementary Figures S7 G, H).
Discussions
A recent study estimated that the hazard ratio of CNS impairment was 1.42 compared to those patients did not require hospitalization during COVID-19 infection [15]. Compared with the healthy control group, the risk rate of neurologic symptoms was obvious in those who were not hospitalized with COVID-19 and increased in patients from non-hospitalized at 7% to hospitalized and to ICU, with the rate increasing to 12% [15].
A recent study reported that neuropathology, observed in most brains from patients with COVID-19, does not directly result from viral infection in the brain, but more likely from systemic inflammation, specifically from hypoxia/ischemia [16]. This is quite consistent with our study, in which the hypoxia score is greatly increased in both stroke and severe COVID patients. Hypoxia is a common pathophysiology in both stroke and COVID. Interestingly, a recent study found that hypoxia had an impact on B cells in COVID infection, including reduced marginal zone-like, memory and transitional B cells [17]. Another study identified cognitive disorders in patients with COVID-19; 18% had memory impairment and 11% developed attentional dysfunction [16]. These studies indicate that hypoxia is associated with the immune status and neurological outcomes in COVID-19. Consistently, involvement of complement has been revealed in the pathogenesis of patients with COVID-19 [18–20]. A previous study showed that the COVID-19 spike protein can activate the complement pathway and cause complement-mediated cell death [18]. The authors summarize that complement dysregulation contributes to the pathogenesis of COVID-19 and might be a marker of disease severity.
Our current bioinformatical study in both COVID-19 and stroke identified several shared gene signatures and their relationship with immune infiltration and metabolic pathways. Among them, GRB10 and CCR5, mainly expressed in monocytes, might be involved in the immune metabolic reprograming in both diseases. The increased values of GRB10 and reduced expression level of CCR5 could reflect subclinical brain involvement in the context of a multisystem disease, such as COVID-19, since the above biomarkers are representative of different elements of central nervous system injury. We recently found that GRB10 was linked to immune deficiency in traumatic brain injury, which is another common form of brain insult [21]. The expression level of GRB10 increased with the severity of brain injury and, accordingly, the NK cell count decreased. In this study, we also found a negative relationship between GRB10 and infiltrated NK cells. Moreover, the CIBERSORT results indicated that GRB10 was also negatively associated with T cells, while CCR5 was positively correlated with CD8 T cells. The exact role of these genes in the regulations of immune cells needs to be validated by experimental study. Furthermore, the significant correlation of GRB10 and CCR5 with immune cell markers, as demonstrated in the present study, is in line with that of brain insult biomarkers with inflammatory chemokine expression from recent research [22–24].
In conclusion, our research combined the analysis results of bulk RNA-seq and scRNA-seq data revealing stroke-related hub genes and their role in the immune and inflammation phenotype in COVID, which can provide useful biomarkers and an interventional strategy.