Introduction
As a type of frequently diagnosed malignancy, gastric cancer (GC) is currently the second leading cause of cancer-related mortality worldwide [1]. In addition, the stages of gastric adenocarcinoma can consist of dysplasia, intestinal metaplasia, atrophic gastritis and Helicobacter pylori infection [2]. So far, endoscopic mucosal resection (EMR) remains a universal and well-established approach in the removal of intra-mucosal malignancy and benign lesions from the gastrointestinal tract. From this perspective, multiple EMR techniques have been developed for clinical applications [3–6]. However, the primary limitation of current EMR techniques is the high post-operative recurrence rate [7]. For example, some studies have reported a recurrence rate of up to 3% in post-EMR patients [8, 9]. And it is noteworthy that in a study by Tao et al., which compared endoscopic submucosal dissection (ESD) with EMR for early stage GC, despite its increased perforation risk and operation time, ESD was proved to exhibit a superior efficacy for en bloc resection, complete resection, curative resection and local recurrence [10]. In addition, the risk of local relapse can be affected by factors such as the number of resections, the location of tumor foci and the presence of residual cancer cells near the site of resection [11].
It was hypothesized that up to 98% of the human genome is occupied by non-coding transcripts. In fact, the complexity in gene expression profiles of individuals may originate from the mutations in these non-coding transcripts [12]. Furthermore, many non-coding transcripts, including long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), have been shown to act as key modulators of gene expression in both pathological and physiological conditions by interacting with their target RNAs, DNAs and proteins [13]. For example, miR-135b-5p was inferred to have a tumor-promoting role in GC cells via down-regulating KLF4, which accordingly suggested that the inhibition of miR-135b-5p could be utilized in the treatment of gastric cancer [14], and lncRNA TP73-AS1 was proved to promote cell proliferation of NSCLC, indicating its potential as a therapeutic target in NSCLC [15].
Taurine up-regulated gene 1 (TUG1), an lncRNA also referred to as ncRNA00080, linc00080 or TI-227H, was initially identified as a RNA transcript whose expression is elevated by taurine. In addition, the inhibition of TUG1 in mice blocked their retinal development [16]. Furthermore, TUG1 expression in human tissues and human cell lines has also been studied, although the role of TUG1 in cancer development remains elusive. Nevertheless, TUG1 was found to regulate cell cycle progression by interacting with polycomb repressive complex 2 (PRC2) [17]. Also, a previous report indicated that TUG1 functions as an oncogenic lncRNA to promote tumor progression by competing as an endogenous ‘sponge’ for miR-382, thus regulating the expression of EZH2 [18].
As an adhesion molecule involved in a wide range of signaling pathways, CD44 can be spliced into various isoforms [19]. For example, the expression of CD44 is universal in most basal cells of normal prostate tissues. However, the expression of CD44 was shown to become aberrant in prostate cancer. For example, a study of 109 prostate cancer patients demonstrated the down-regulation of all CD44 isoforms in up to 98% of the patients [20]. In addition, CD44 was previously shown to act as an anti-apoptotic factor to promote the metastasis and invasion of multiple cancers, while the increased expression of CD44 was implicated in the relapse of GC. In another study [21], the expression of CD44 protein and mRNA was evidently higher while the expression of miR-328 was significantly decreased in a group of GC patients suffering from cancer relapse, suggesting the presence of a negative regulatory relationship between CD44 and miR-328 in patients with recurrent GC. In addition, it was shown that the inhibition of HIF-1α expression in GC cells inhibited the expression of CD44 and suppressed the invasiveness and growth of GC cancer cells. These results suggested that the expression of CD44 is regulated by HIF-1α, thus affecting the growth and metastasis of GC [21]. And it is also noteworthy that for GC pathogenesis, high percentages of lymphocytes with CD200 expression may contribute to the development of chronic inflammation and influence gastric carcinogenesis [22].
It has been reported that miR-382 could act as a biomarker to predict post-ESD relapse of GC [23]. In addition, as a target of miR-382, CD44 was shown to regulate the proliferation of GC cells [23]. Furthermore, TUG1 was found to function as an endogenous competing RNA to inhibit the effect of miR-382 [24]. In this study, we investigated the prognostic value of TUG1 in the prediction of post-EMR GC relapse.
Material and methods
Human subjects and sample collection
In this study, we recruited 112 early stage GC patients who were diagnosed with early stage GC (early gastric cancer (EGC) is defined as invasive gastric cancer that invades no more deeply than the submucosa) by white light endoscopy and chromoendoscopy. These patients, ranging in age from 46 to 63 years and with an approximate sex ratio of 11 : 10 (male patients – 59, female patients – 53), received EMR treatments to remove part of their stomach during the period from January 2013 to April 2015 at Chinese PLA Hospital No. 254. The collected information about the subjects enrolled in this study is presented in Table I. The issue specimen obtained after EMR and peripheral blood samples were collected from each subject and stored at –80°C until used for further investigation. Patients were further grouped according to the median level of TUG1 of their TUG1 expressions in blood and tissue samples. The protocols of the present study were approved by the Institutional Ethics Committee. Written forms of informed consent were signed and obtained from all participating patients.
Table I
Collected information about the subjects enrolled in this study
RNA isolation and real-time PCR
The mRNA expression of TUG1, miR-382 and CD44 in collected tissue/blood samples and cultured cells was measured using real-time PCR. The total RNA was extracted by a Trizol kit (Invitrogen, Carlsbad, CA), with the concentration and purity of isolated RNA being determined by a spectrophotometer. The sample RNA was reversely transcribed into cDNA using a Primescript TMR Reverse Transcription Kit (TaKaRa, Tokyo, Japan), and the cDNA product from each sample was diluted with 65 µl of diethyl phosphorocyanidate (DEPC) water and thoroughly mixed, and the mRNA expression of TUG1, miR-382 and CD44 in each sample was measured by real-time PCR using a SYBR Premix Ex Taq II kit (TaKaRa, Tokyo, Japan) in conjunction with an ABI PRISM 7300 system (ABI, Foster City, CA). The reaction cycles of real-time PCR included 4 min of pre-denaturation at 95°C, followed by a total of 40 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min. At the end of qPCR, a 7-min extension cycle was performed at 72°C. During qPCR, GAPDH and U6 were used as the internal references. The relative expression of TUG1 (forward primer: 5’-GCAAACTGAGGATGCTCCATCC-3’; reverse primer: 5’-TACCAGGTCTGTAGGCTGATGG-3’), miR-382 (forward primer: 5’-GTTGTTCGTGGTGGATTC-3’; reverse primer: 5’-GAACATGTCTGCGTATCTC-3’) and CD44 (forward primer: 5’-CCAGAAGGAACAGTGGTTTGGC-3’; reverse primer: 5’-ACTGTCCTCTGGGCTTGGTGTT-3’) was calculated using the 2–ΔΔCt method. Each experiment was repeated three times.
Cell culture and transfection
AGS and HGC-27 cells were cultured in a RPMI 1640 culture medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin. The cells were cultured in an incubator under saturated humidity, 5% CO2 and 37°C. Sub-culture was conducted when cell confluence reached 80–90%. For transfection, the cells were seeded into 24-well plates at a density of 2 × 105 cells per well and cultured overnight. Subsequently, the cells were transfected with pcDNA-TUG1 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer’s protocol. The cells were harvested at 48 h after transfection for subsequent experiments.
Vector construction, mutagenesis and luciferase assay
By informatics analysis of potential target genes of miR-382, a miR-382 binding site was found in TUG1 as well as in the mRNA of CD44. The full length of TUG1 and CD44 mRNA was amplified and cloned into pcDNA3.1 vectors (Promega, Madison, WI). Meanwhile, site-directed mutagenesis was carried out in the miR-382 binding sites of TUG1 and CD44 mRNA, which were later amplified and cloned into pcDNA vectors. Subsequently, Lipofectamine 2000 was used to co-transfect the AGS and HGC-27 cells with miR-382 and mutant/wild type TUG1 or CD44 plasmids. The cells were harvested 48 h later. Luciferase activity of transfected cells was detected using a dual-luciferase reporter gene detection assay (Promega, Madison, WI) on a bioluminescence detector (Modulus, Promega, Madison, WI).
Cell proliferation assay
After AGS and HGC-27 cells, which were obtained from ATCC, Manassas, VA, were transfected for 48 h in a 96-well plate, 20 µl of MTT reagent (Sigma-Aldrich, St. Louis, MO; diluted by PBS to 5 mg/ml) was added to each well after the cells were incubated with DMEM media containing 10% FBS and 5% CO2 at 37oC for 12, 24 and 48 h. After discarding the supernatant, 150 µl of dimethyl sulfoxide (DMSO, Sigma-Aldrich, St. Louis, MO) was added to each well to dissolve the formazan crystals. The optical densities (OD) in the wells were measured at 490 nm on a microplate reader in order to calculate cell viability.
Western blot analysis
Total protein was extracted from tissue and cell samples using an RIPA lysis buffer (Solarbio Science, Beijing, China) containing phenylmethanesulfonyl fluoride (PMSF). Following 30-minute incubation on ice, the samples were centrifuged at 12,000 r/min and 4°C for 10 min to collect the supernatant, whose total protein concentration was then evaluated using a bicinchoninic acid (BCA) protein kit (Pierce, Rockford, IL). 50 µg of protein from each sample was resolved using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, Beyotime Biotechnology, Shanghai, China) and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were then blocked with Tris-buffered saline-Tween-20 (TBST) containing 5% skimmed milk for 2 h, followed by incubation at 4°C overnight with anti-CD44 primary antibodies (1 : 1000, Abcam, Cambridge, MA). Subsequently, the membranes were rinsed with PBS and incubated with horseradish peroxidase (HRP)-conjugated IgG secondary antibodies (1 : 2000, Abcam, Cambridge, MA) at room temperature for 2 h. Finally, the membranes were developed using an enhanced chemiluminescence (ECL) reagent (Millipore, Billerica, MA). The relative protein expression of CD44 was determined based on the ratio between the gray value of the CD44 protein band and the grey value of the β-actin (internal reference) protein band.
Apoptosis analysis
The apoptotic profiles of cell samples were measured by flow cytometry using an Annexin V-FITC apoptosis detection kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions.
Statistical analysis
All data analyses were conducted using SPSS 22.0 software (IBM, Armonk, NY). All measurement data were expressed as mean ± standard deviation (SD). The analysis of relapse-free survival curves was carried out using the log rank test module in Biostar software. The differences between groups with high and low TUG1 expression, as well as the differences between the TUG1-transfected group and control group, were compared using t-tests, while the differences among multiple groups in the luciferase assay were compared using one-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparison test as the post hoc test. A p-value of < 0.05 was considered as statistically significant.
Results
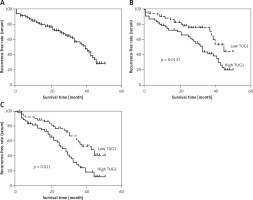
Diagnostic value of TUG1 in predicting post-EMR relapse of GC
In this study, we used the median level of TUG1 as an indicator to divide the patients into a high TUG1 expression group and a low TUG1 expression group. Interestingly, no significant difference was observed between these two groups in terms of patient characteristics, including age, sex, BMI, the presence of H. pylori infection, and the history of alcohol abuse and smoking (Table I). The relapse-free rate of all patients was analyzed for up to 60 months using the log rank test (Figure 1 A). Moreover, the AUC of the high TUG1 expression group was obviously smaller than that of the low TUG1 expression group for both blood (Figure 1 B) and tissue samples (Figure 1 C). These results indicated that the level of TUG1 could be used as a biomarker to predict the rate of post-EMR GC relapse.
Figure 1
AUC of the high TUG1 expression group and low TUG1 expression group for both blood and tissue samples. A – AUC of relapse-free survival of up to 60 months in the blood sample of all patients recruited. B – AUC of relapse-free survival of up to 60 months in the blood sample groups with high and low TUG1 expression. C – AUC of relapse-free survival of up to 60 months in the tissue sample groups with high and low TUG1 expression
Differentiated expression of miR-382 and CD44 in patients with different levels of TUG1 expression
Subsequently, we measured the relative expression of TUG1 and miR-382, as well as the mRNA/protein levels of CD44, in all blood and tissue samples. As shown in Figure 2 A, the relative expression of TUG1 was different in different groups of samples. Interestingly, the relative expression of miR-382 (Figure 2 B) was apparently up-regulated in the tissue group with a lower level of TUG1 expression, along with decreased mRNA (Figure 2 C) and protein (Figure 2 D) expression of CD44. Similar results were also observed in blood samples (Figure 3), indicating the presence of a negative relationship between the expression of miR-382 and its endogenous competing RNA TUG1, as well as between the expression of miR-382 and its target gene CD44.
Figure 2
Correlation between TUG1, miR-382 and CD44 mRNA/protein expression in the tissue sample groups with high and low TUG1 expression (*p < 0.05 vs. tissue sample group with high TUG1 expression). A – Relative expression of TUG1 in the tissue sample groups with high and low TUG1 expression. B – Relative expression of miR-382 in the tissue sample groups with high and low TUG1 expression. C – Relative expression of CD44 mRNA in the tissue sample groups with high and low TUG1 expression. D – Relative density of CD44 protein in the tissue sample groups with high and low TUG1 expression
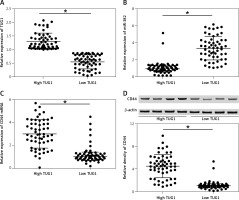
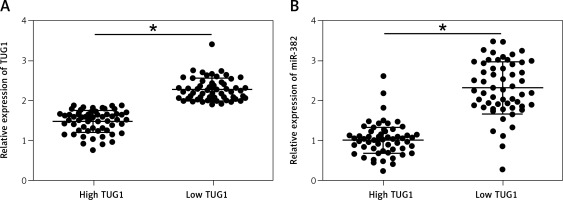
Figure 3
Correlation between TUG1, miR-382 and CD44 mRNA/protein expression in the blood sample groups with high and low TUG1 expression (*p < 0.05 vs. tissue sample group with high TUG1 expression). A – Relative expression of TUG1 in the blood sample groups with high and low TUG1 expression. B – Relative expression of miR-382 in the blood sample groups with high and low TUG1 expression
Establishment of a TUG1/miR-382/CD44 signaling pathway
As demonstrated by the results obtained from the luciferase assay, the luciferase activity of AGS (Figure 4 A) and HGC-27 cells (Figure 4 B) was significantly reduced after the co-transfection with wild-type TUG1 and miR-382, while the luciferase activity of AGS (Figure 4 A) and HGC-27 cells (Figure 4 B) co-transfected with mutant TUG1 and miR-382 showed no significant difference. Moreover, the luciferase activity of AGS (Figure 4 C) and HGC-27 cells (Figure 4 D) was also significantly reduced after the co-transfection with wild-type CD44 and miR-382, while the luciferase activity of AGS (Figure 4 C) and HGC-27 cells (Figure 4 D) co-transfected with mutant CD44 and miR-382 showed no significant difference. Therefore, a TUG1/miR-382/CD44 signaling pathway could be established. In addition, TUG1 negatively regulated the expression of miR-382, which in turn negatively regulated the expression of CD44.
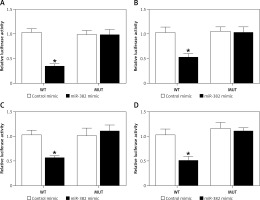
Figure 4
Establishment of a TUG1/miR-382/CD44 signaling pathway (*p < 0.05 vs. cells transfected with control microRNAs). A – Luciferase activities of AGS cells co-transfected with miR-382 mimics/control microRNAs and plasmids carrying the wild-type/mutant TUG1. B – Luciferase activities of HGC-27 cells co-transfected with miR-382 mimics/control microRNAs and plasmids carrying the wild-type/mutant TUG1. C – Luciferase activities of AGS cells co-transfected with miR-382 mimics/control microRNAs and plasmids carrying the wild-type/mutant CD44 mRNA. D – Luciferase activities of HGC-27 cells co-transfected with miR-382 mimics/control microRNAs and plasmids carrying the wild-type/mutant CD44 mRNA
Effect of the TUG1/miR-382/CD44 signaling pathway on cell proliferation
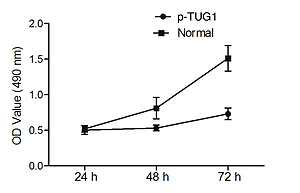
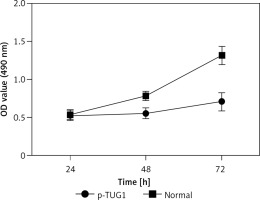
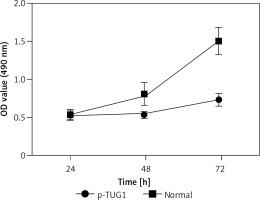
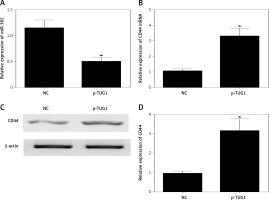
The regulatory relationship involved in the TUG1/miR-382/CD44 signaling pathway was evaluated by measuring the effect of TUG1 on the relative expression of miR-382 and CD44 mRNA/protein. As demonstrated in Figure 5, the relative expression of miR-382 (Figure 5 A) was obviously inhibited in AGS cells transfected with TUG1, while the relative expression of CD44 mRNA was evidently up-regulated after TUG1 transfection. Similarly, the expression of CD44 protein was also increased in AGS cells transfected with TUG1. Furthermore, the transfection of TUG1 into AGS cells (Figure 6) significantly promoted cell proliferation, as indicated by the increased OD values detected via the MTT assay. When the above experiments were repeated in HGC-27 cells (Figures 7 and 8), similar results were obtained, confirming the role of TUG1 as an enhancer of both CD44 expression and cell proliferation via down-regulating the expression of miR-382. Therefore, the reduced expression of TUG1 led to reduced expression of CD44, which in turn inhibited GC proliferation and lowered the risk of post-EMR GC relapse.
Figure 5
Effects of TUG1 on expression levels of miR-382 and CD44 in AGS cells (*p < 0.05 vs. negative control group). A – Relative expression of miR-382 in AGS cells in the presence or absence of TUG1. B – Relative expression of CD44 mRNA in AGS cells in the presence or absence of TUG1. C – Intensity of CD44 protein band in AGS cells in the presence or absence of TUG1. D – Relative density of CD44 protein in AGS cells in the presence or absence of TUG1
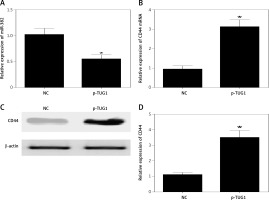
Figure 7
Effects of TUG1 on expression levels of miR-382 and CD44 in HGC-27 cells (*p < 0.05 vs. negative control group). A – Relative expression of miR-382 in HGC-27 cells in the presence or absence of TUG1. B – Relative expression of CD44 mRNA in HGC-27 cells in the presence or absence of TUG1. C – Intensity of CD44 protein band in HGC-27 cells in the presence or absence of TUG1. D – Relative density of CD44 protein in HGC-27 cells in the presence or absence of TUG1
Discussion
Although endoscopic submucosal dissection (ESD) is now applied as a novel treatment for endoscopic removal of early stage gastric cancer, EMR is utilized as a conventional approach in the treatment of early stage GCs [25]. In this study, all samples were collected after EMR and all results were referring to survival after EMR only. The residual tumor cells overlooked by EMR during surgical resection often lead to tumor relapse, thus reducing the efficacy of EMR treatment. In fact, up to 18% of patients treated with EMR ultimately succumbed to local relapse [26, 27]. It has been shown that the expression of lncRNA TUG1 is high in GC tissues. Further analyses based on a more defined classification of GC subtypes showed that the expression of TUG1 in GC patients with different clinicopathological characteristics was closely related to the status of lymphatic metastasis and the TNM staging. A previous study indicated that lncRNA-TUG1 was up-regulated in GC tissues, which also promoted the invading capacity of GC by inhibiting the pathway of microRNA-144/c-Met [28]. The results of several recent studies also demonstrated a similar role of TUG1 in glioma and osteosarcoma [29–31]. Furthermore, Transwell assays and scratch tests were used to investigate the biological role of TUG1 during cancer pathogenesis by inhibiting the expression of TUG1 in SGC-7901 cells using siRNA. The results of the above assays showed that the silencing of TUG1 could suppress the invasiveness and metastasis capability of SGC-7901 cells. In addition, both cytological and histological experiments demonstrated that TUG1 played a certain role in promoting the metastasis of GC cells. In this study, blood and tissue samples were collected from 112 early stage GC patients undergoing EMR treatments. Subsequently, the samples were grouped according to the level of TUG1 expression in the patients. We found that the relapse-free survival of patients in the high TUG1 expression group was longer than that in the low TUG1 expression group, indicating that the level of TUG1 could be used to predict the risk of post-EMR GC relapse.
MiR-328 has been implicated in the relapse of early stage GC patients undergoing EMR treatments. In one study, 230 early stage GC patients were enrolled to undergo EMR treatments and their relapse-free survival was evaluated using a Cox proportional hazard model. The results showed that, compared to that in patients showing high expression of miR-328, the duration of relapse-free survival was significantly lower in patients with lower miR-328 expression, indicating an important role of miR-328 in the susceptibility to cancer relapse. Meanwhile, another study also indicated that the down-regulation of miR-382 expression was associated with the progression of gastric cancer, while the up-regulation of miR-382 expression could inhibit the migration, invasion and proliferation of MGC-803 cells [32]. It was also shown that miR-328 could target CD44 and suppress its expression, whereas the suppression of miR-328 in the tumor microenvironment may increase the expression of CD44 on macrophages, eventually inducing ROS defense and promoting tumor progression [33]. Furthermore, it was shown that the de novo expression of CD44 in gastric mucosa was closely related to the induction of chronic inflammation and the downregulation of miR-328 expression in gastric mucosa infected by H. pylori virus, suggesting the involvement of CD44-mediated miR-328 silencing in the pathogenesis of GC. A previous report indicated that the expression of CD44 and its splice variants was positively associated with the initiation and progression of gastric cancer, thus implicating CD44 in the diagnosis, treatment and prognosis of gastric cancer [34]. In this study, we also confirmed that miR-382 was highly expressed in samples with low TUG1 expression, while the expression of CD44 mRNA/protein showed a similar trend as that of TUG1. Therefore, a negative correlation was established between the expression of miR-382 and TUG1, as well as between the expression of miR-382 and its target gene CD44.
As an important molecule expressed in the extracellular matrix, CD44 has been implicated in many processes of tumorigenesis, including the invasion and metastasis of tumor cells [35]. In addition, CD44 has been shown to act as a cell membrane marker in a wide range of solid tumors [36–38]. In a previous study, it was demonstrated that a mutation in CD44 could alter the risk of post-EMR GC relapse [34]. In addition, CD44 was previously identified as a target of miR-328 in both normal mucosal cells and cancer cells [33, 39]. Moreover, the overexpression of CD44 has been detected in both primary cancer tissues and cultured cancer cell lines, and the expression of CD44 was shown to promote the proliferation of cancer cells [40–42]. Some scholars have hypothesized that the overexpression of miR-328 in GC cells could inhibit cancer cell proliferation by downregulating the expression of CD44. Other researchers have carried out proliferation assays which showed that the growth of HCT116 and KATOIII cells was significantly inhibited by transfecting the cells with miR-328. Furthermore, the knockdown of miR-328 expression in cancer cells not only increased CD44 expression, but also enhanced the proliferation of these cancer cells. In this study, TUG1 was shown to inhibit the expression of miR-382 while increasing the expression of CD44 mRNA/protein. In addition, the rate of cell proliferation was evidently elevated by TUG1. Therefore, TUG1 is implicated in the pathogenesis of GC by promoting the proliferation of GC cells and by increasing the expression of CD44, thus leading to an increased risk of post-EMR GC relapse. Another recent study also showed that the expression of CD44 was associated with the magnitude of distant metastasis and lymph node metastasis. In addition, a higher level of CD44 expression also correlated with a higher TNM grade and a poorer prognosis in GC patients [43]. Finally, as a glycoprotein receptor present on the cell surface that affects the capability of cell adhesion, CD44 was shown to directly regulate the metastasis and migration potential of GC cells [44]. As one of the most important cell adhesion molecules, CD44 has been shown to participate in the survival, proliferation, matrix degradation, extracellular matrix adhesion, and motility of cancer cells [17, 45, 46]. Therefore, CD44 plays indispensable roles in cancer pathology, such as tumor metastasis and progression. It has also been reported that the downregulation of CD44 in GC cells could affect the invasion and proliferation potential of these cells, thus highlighting the prospect of using CD44 as a therapeutic target in GC treatment.
In conclusion, the findings of this study demonstrated that miR-328 could act as a biomarker to predict post-EMR GC relapse. By targeting the expression of CD44, miR-328 was also shown to regulate the proliferation of GC cells. In addition, TUG1 was found to function as an endogenous competing RNA to inhibit the effect of miR-328. Therefore, TUG1 could also be used as a biomarker to predict post-EMR GC relapse.