MiRNA-21 is abnormally expressed in many cardiovascular diseases [1]. Recently, it has been reported that miRNA-21 is involved in myocardial protection. Injection of exogenous synthetic miRNA-21 can reduce the size of myocardial infarction by 64% [2]. Further microarray analysis shows that many apoptosis related genes could not be inhibited by miRNAs. MiRNA-21 is involved in the proliferation, apoptosis and differentiation of a variety of cells, including bone marrow mesenchymal stem cells (BMSCs).
In this study, we found that miR-21 could regulate the expression of ajuba by targeting its 3′UTR. MiR-21 overexpression promoted the expression of Isl1 at both transcriptional and translational levels. Ajuba downregulation increased the expression of Isl1 and promoted differentiation of BMSCs into cardiomyocyte-like cells, which could be a therapeutic target for many cardiovascular diseases.
Methods
Isolation, culture, induction, and identification of characteristic antigens of BMSCs were described in a previous article [3]. For prediction of miRNA-21 target genes we used mirwalk software to analyze the interaction of all candidate target genes through TargetScan, mirdb and Miranda.
In construction of the miRNA-21 expression virus vector (mir-21-OE), the steps are the same as in the article by Yang et al. [3]. The MiRNA-21 sponge (mir-21-KD) contains the target miRNA-21 complementary binding site. According to the miRNA-21 sequence, the miRBase access No. is mi0000850. The primer sequence of the miR-21 sponge is as follows: miR-21 sponge f(XhoI): ccgctcgagctcgtcaacatcagtctg and miR-21 sponge R (BamHI): ccggatccttggtgagcttatcagac. Specific operation steps were according to the manufacturer’s instructions.
Construction and virus packaging of the ajuba overexpression virus vector (ajuba-OE) and ajuba interference virus vector (ajuba-KD): ajuba sequence: nm according to access No_ 053503.1. The plasmid template was purchased from Beijing Yiqiao Shenzhou Biotechnology Co., Ltd. Through sub-cloning, the relevant primer sequences are as follows: ajub-f (xhoi): ccgctcgagatggtggttaggggagaaag; and ajub-r (BamHI): ccgggatcctcagatatagttggtagggggctg.
For construction of the shRNA lentivirus interference vector we used plko.1-gpf-puro vector through the BLOCK-iT RNAi designer tool (https://rnaidesigner.thermofisher.com/rnaiexpress/). The designed shRNA was synthesized by Shanghai Shenggong Bioengineering Co., Ltd.
The RT-PCR, Western blotting, and immunofluorescence staining detection of BMSCs in each group were all according to the manufacturer’s instructions. Luciferase reporter assays in BMSCs: The wild-type 3′UTR of ajuba (ajuba-3′UTR-WT) and mutant 3′UTR of ajuba (ajuba-3′UTR-MUT) expression vectors were constructed by gene synthesis. BMSCs were cultured and inoculated in 24-well plates for 10–24 h (80% confluence). BMSCs were infected with the virus with miRNA-21-OE and miRNA-21-KD, and then the reporter gene plasmid was co-transfected into the cells.
Results
By bioinformatics prediction, ajuba was predicted as a potential target gene of miRNA-21 in BMSCs, as shown in Figure 1 G: BMSC culture and cell characterization were according to the method’s of Yang et al. [3]. miRNA-21-KD was successfully constructed by gene synthesis, as shown in Figure 1 A. The inhibition rate of miR-21 detected by RT-qPCR was 75% (p < 0.001), as shown in Figure 1 B. Ajuba-OE was successfully constructed by the subclonal method, as shown in Figure 1 C. The overexpression of ajuba OE detected by RT qPCR was 75% (p < 0.001), as shown in Figure 1 D. Ajuba-KD was constructed, and the shRNA sequence was confirmed to be completely correct by sequencing, as shown in Figure 1 E. The expression of ajuba at the protein level was detected by WB. Shrna-3 can be used to interfere with lentivirus in subsequent functional experiments, as shown in Figure 1 F.
Figure 1
A – Results of digestion and identification of miRNA-21 sponge vector:Xhoi and bamh1 were digested and identified as 190 bp, which was consistent with the expected size. Lane 1: pcdh empty; Lane 2: xhoi and bamh1 digestion identification; Lane M: DNA marker. B – MiR-21 sponge significantly inhibits miR-21 expression in BMSC cells. C – Results of enzyme digestion of ajuba OE vector, Lane 1: pcdh empty; Lane 2: xhoi and bamh1 digestion identification; Lane M: DNA marker. D – Detection of overexpression of BMSC infected by ajuba OE lentivirus A, a – The expression of ajuba mRNA was significantly increased by RT qPCR, b – The expression of ajuba protein was detected by WB, and the expression of ajuba increased. E – Results of enzyme digestion of ajuba OE vector, Lane 1: plko. 1 empty; Lane 2: EcoRI digestion identification; Lane M: DNA marker.Compared with shRNA NC control virus, the mRNA expression of ajuba was detected by RT qPCR. The results showed that the inhibition rates of ajuba shrna-1, ajuba shrna-2 and ajuba shrna-3 were 35.12%, 74.35% and 84.35% (*p < 0.05). F – Overexpression detection of BMSC infected by ajuba shRNA lentivirus, a – The expression of ajuba mRNA was significantly inhibited by RT qPCR, b – The expression of ajuba protein was detected by WB, and ajuba was significantly inhibited. β-actin is an internal parameter. ***p < 0.001. G – The prediction binding site between miRNA-21 and the 3’UTR of Ajuba
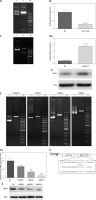
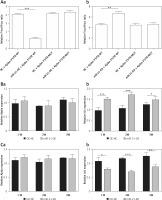
Figure 2 shows that mir-21-OE and mir-21-KD infected BMSCs. The luciferase reporter gene detected the direct regulatory effect of miR-21 on ajuba. MiRNA-21 regulates the expression of Isl1 both at transcriptional and translational levels in BMSCs. In the mir-21-OE group, the expression of Isl1 was significantly higher than that of the control group. In the mir-21-KD group, the expression of Isl1 was significantly lower than in the control group.
Figure 2
A – Detection results of dual luciferase reporter gene, a – Compared with kd-nc group, the activity of reporter gene transfected with wild-type expression vector in mir-21-kd group was significantly higher (p < 0.001); It can be seen that by inhibiting miR-21, the expression of wild-type ajuba protein is inhibited, so the expression is significantly high, b – Wt: wild type ajuba 3’UTR; Mut: mutant ajuba 3’UTR, **p < 0.01, ***p < 0.001. B – Detection of ajuba and Isl1 mRNA expression in BMSC during mir-21-oe lentivirus infection and differentiation, a – There was no significantd ifference in ajuba mRNA expression at 1 week, 2 weeks and 3 weeks by RT qPCR, b – The expression of Isl1 mRNA was significantly increased at 1 week, 2 weeks and 3 weeks by RT qPCR. β-actin is an internal parameter. ***p < 0.001, *p < 0.05. C – Detection of ajuba and Isl1 mRNA expression in BMSC during mir-21-kd lentivirus infection and differentiation, a – There was no signific ant difference in ajuba mRNA expression at 1 week, 2 weeks and 3 weeks by RT qPCR, b – The expression of Isl1 mRNA decreased significantly at 1 week, 2 weeks and 3 weeks by RT qPCR, β-actin is an internal parameter. ***p < 0.001, **p < 0.01, *p < 0.05
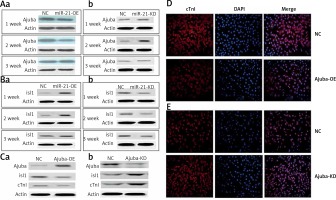
We found that when the expression of ajuba protein decreased, the expression of Isl1 protein was increased as detected by WB in the mir-21-OE group compared with the control group, and vice versa, as shown in Figures 3 A, B. After ajuba-OE was used, when the expression levels of ajuba increased, the expression of Isl1 and cTnI decreased, and vice versa, as shown in Figure 3 C. As shown in Figure 3 D, with over-expression of ajuba, the expression of cTnI protein was decreased as detected by immunofluorescence, compared with the control group, and vice versa.
Figure 3
A – Detection of ajuba protein expression in BMSC during lentiviral infection and differentiation of mir-21-OE and mir-21-KD, a – WB was used to detect the expression changes of ajuba in 1 week, 2 weeks and 3 weeks after miR-21 over-expression, b – After the decrease of miR-21 was detected by WB. B – Detection of Isl1 protein expression in BMSC during lentiviral infection and differentiation of mir-21-OE and mir-21-KD, a – WB was used to detect the expression of Isl1 in 1 week, 2 weeks and 3 weeks after miR-21 overexpression, b – WB was used to detect the changes of Isl1 expression in 1 week, 2 weeks and 3 weeks after the decrease of miR-21. C – Detection of ajuba, Isl1 and cTnI protein expression in BMSC after ajuba overexpression and ajuba interference, a – WB was used to detect the expression changes of ajuba, Isl1 and cTnI proteins after ajuba overexpression, b – WB was used to detect the expression changes of ajuba, Isl1 and cTnI proteins after ajuba interference knockdown. D – After ajuba overexpression, the expression of cTnI protein was detectedby immunofluorescence. E – The expression of cTnI protein in ajuba interference knockdown group and control group was detected by immunofluorescence. The photo magnification is 200×
Discussion
MiRNA-21 is involved in the proliferation, apoptosis and differentiation of a variety of cells [4, 5], and related target genes and possible related pathways are reported [6], However, the mechanism is not clear in the cardiovascular system. In order to improve the role of BMSCs in the treatment of ischemic diseases, especially coronary heart disease, it is necessary to carry out further exploration.
Ajuba contributes to the formation and maintenance of cell-cell connections and plays a role in cell division and cell migration. Ajuba is generally combined with transmembrane proteins or signal proteins to mediate a wide range of biological functions, including cell proliferation and apoptosis and cell differentiation. Some studies have also shown that ajuba protein is involved in adipocyte differentiation [7, 8]. In our study, ajuba was predicted as a target gene whose expression could be regulated by miRNA-21, and our data also proved that miRNA-21 negatively regulated the expression of ajuba in BMSCs. Isl1 is a pioneer factor in cardiomyocytes. The latest research shows that BMSCs transfected with the Isl1 gene can significantly promote the differentiation of bone marrow mesenchymal stem cells into cardiomyocyte like cells [9]. In this experiment, it was found that the expression of Isl1 increased during the induced differentiation of BMSC cells. Similarly, when the expression of ajuba increased, the expression of Isl1 was inhibited, showing the decline of the ability of BMSC cells to induce differentiation. We preliminarily proposed to upregulate miRNA-21, target inhibit ajuba expression, and enhance Isl1 expression to promote BMSC differentiation; that is, miRNA-21 regulates BMSC differentiation through targeting the Isl1 axis, and affects the differentiation of bone marrow mesenchymal stem cells into cardiac muscle like cells, which may play a role in the regeneration and differentiation of bone marrow mesenchymal stem cells and the improvement of cardiac function by BMSC transplantation in the treatment of heart diseases. In conclusion, we observed that miRNA-21 regulated the expression of Isl1 through targeting ajuba in BMSCs, which might be used to regulate the differentiation of BMSCs to cardiomyocyte-like cells in clinical application.