Introduction
Liver cancer can be roughly categorized into four subtypes – hepatocellular carcinoma (HCC), hepatoblastoma, cholangiocarcinoma and angiosarcoma. Hepatocellular carcinoma is the most common and prevalent cancer type among them [1]. In the last decades, HCC has been listed as one of the five most common malignancies worldwide [2]. In 2015, about 4.3 million patients suffered from HCC and 2.8 million people died of the disease in China [3]. Approximately 70% of HCC patients who underwent liver transplantation or surgical resection still faced a higher risk of recurrence in 5 years [4]. In addition, the 5-year overall survival rate of HCC is considerably low, which forebodes a poor prognosis [5]. This condition prompted researchers around the world to focus on the mechanism of HCC and identify new clinical treatments for the patients. Some biomolecules have been confirmed to play vital roles in the progression of HCC, such as adrenomedullin [6] and cyclooxygenase-2 (COX-2) [7]. In recent years, the co-relationship between microRNAs and their target genes, and its effect on the carcinoma cells, has become a research hotspot, and HCC is no exception.
MicroRNA (miRNA) is a kind of endogenous small RNA with a length of about 20 to 24 nucleotides. MiRNAs can function efficiently in the pathogenesis of many malignant carcinomas as either an oncogene or tumor suppressor [8–10]. For example, Zhao et al. found that miR-630 functioned as an oncogene in renal cell carcinoma [11]. Wang et al. found that the dysregulation of miRNA-222 exerted an influence on the metastasis and invasion of thyroid cancer cells [12]. Li and Luo revealed the possible acceleration of nasopharyngeal carcinoma by miR-27a-3p overexpression [13]. The aberrant expression of miRNA will also influence the progression of HCC. Li et al. demonstrated that miR-146b-5p could inhibit migration, proliferation, invasion and induced cell apoptosis in both vitro and vivo [14]. Ge and Gong reported miR-590-3p involvement in HCC progression and suppression of HCC cells’ growth by targeting the gene TEAD1 [15]. Numerous miRNA- related studies have revealed that the progression of HCC might be related to different miRNAs and their target genes.
After considering previous studies, a widely researched miRNA in many kinds of carcinomas, miR-490, was selected as the topic of our study on HCC. Many studies have reported that miR-490 could influence progression of different tumors though regulating multiple genes. Jia et al. found that miR-490 inhibited the progression of breast cancer through down-regulating the expression of TNKS2 [16]. Chen et al. reported that miR-490 targets the gene PIK3CA to inhibit tumor growth in renal carcinoma cells [17]. Chen et al. reported that miR-490 would suppress ovarian tumorigenesis and progression of epithelial carcinoma by targeting the gene CDK1 [18]. Moreover, Zhang et al. found that miR-490-3p modulated HCC cell growth through targeting ERGIC3 [19], which convinced us that miR-490-3p was of great research potential in HCC progress.
Aurora kinase A (AURKA), a potential target gene of miR-490-3p predicted by the TargetScan database, has been reported as a correlating factor in tumorigenesis and progression. Mignogna et al. reported that AURKA was an oncogene in high-grade serous ovarian carcinoma and overexpression of AURKA led to worse overall survival [20]. Dos Santos et al. found that in lung cancer, AURKA was targeted by mutation of KRAS (kirsten rat sarcoma viral oncogene) and would promote tumor progression [21]. Bornschein et al. also reported that AURKA was associated with gastric cancer and was a negative prognostic indicator of gastric cancer [22]. However, there has been no research focusing on the mechanism of AURKA in HCC, which drew our attention and impelled us to perform the study about miR-490-3p and AURKA, to illuminate their effects on proliferation and migration of HCC cells, and we hope to supply a new theory branch for the clinical prevention and treatment of HCC.
Material and methods
Tissue samples
Hepatocellular carcinoma tissues and adjacent tissues were collected from 78 patients (51 males, 27 females, age: 48–77, average age: 63.5) who underwent surgery in the Affiliated Hospital of Hebei University of Engineering during the period between January 2015 and February 2016. Their tissues were stored in liquid nitrogen. None of the included patients had received any adjuvant treatments, including radiotherapy or chemotherapy. Written informed consent was provided by all patients and this study had been approved by the Ethical Committee of the Affiliated Hospital of Hebei University of Engineering.
Cell culture
Human embryonic kidney cell line HEK-293T, human HCC cell lines Huh-7, HepG2, Hep3B and SK-HEP1 and normal liver cell line HL-7702 were purchased from the Japanese Collection of Research Bioresources (Tokyo, Japan). The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco BRL, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS) (Gibco) and incubated at 37°C in a humidified chamber containing 5% CO2.
Cell transfection
Firstly, AURKA cDNA and pcDNA3.1 plasmid (Promega, Madison, WI, USA) were amplified to construct a lentivirus vector, then reconstruction plasmids were harvested with the Wizard SV96 plasmid DNA purification system (Promega). When cells (HepG2 and Hep3B) reached 80% confluence, Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was applied to transfect miR-490-3p mimics, AURKA siRNA and AURKA cDNA in line with the manufacturer’s guidelines. All reagents were synthesized and purified by Molbase (Shanghai, China). The cells were then cultured and harvested after 24 h transfection.
Quantitative real-time polymerase chain reaction
The total RNA was extracted using Trizol reagent (Gibco). Then complementary DNA (cDNA) was synthesized with the PrimeScript RT reagent kit (TaKaRa, Tokyo, Japan). Expression levels of mRNAs were investigated with the Synergy Brands (SYBR) Premix ExTaq quantitative PCR kit (Thermo Fisher Scientific, Lafayette, Colorado, USA) and Light Cycler instrument (Roche, Basel, Switzerland). U6 expression was then performed as an endogenous control for miR-490-3p normalization, while GAPDH was used for an internal reference for AURKA normalization. The relative expression of miR-490-3p and AURKA was quantified using the 2–ΔΔCt method. Table I shows primers used in quantitative real-time polymerase chain reaction (qRT-PCR), which were synthesized by Shanghai Biotechnology (Shanghai, China).
Table I
Primer sequences designed for quantitative real-time polymerase chain reaction
Western blot
After 24 h transfection, we used radio-immunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China) to extract total protein from tissues or cells. BCA Protein Assay Kit (Vigorous Bio-technology, Beijing, China) was used to concentrate total protein. Proteins were first separated with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Bio-Rad, Hercules, CA, USA) and then transferred to polyvinylidene difluoride (PVDF) membranes (Invitrogen). The membranes were then blocked in 5% non-fat milk for 1 h at room temperature. The primary antibody was rabbit anti-AURKA (1 : 4000, Abcam, Cambridge, MA, USA), and rabbit anti-GAPDH (1 : 2500, Abcam) was used as internal reference. The second antibody was HRP-conjugated goat anti-rabbit antibody IgG (1 : 2000, Abcam). ECF western blot kit (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) was used to reveal the protein bands and Typhoon 9400 imager (GE Healthcare Bio-Sciences) was used to measure the statistics.
Dual-luciferase reporter assay
According to TargetScan (http://www.targetscan.org/), AURKA was one of miR-490-3p’s targets. Sequences containing the miR-490-3p wild-type (wt) and mutant subtype (mut) target region of AURKA 3′UTR (Sangon, Shanghai, China) were inserted into the pmirGLO vector (Promega, USA). The pmirGLO recombinant vector was then co-transfected with miR-490-3p mimics in HEK-293T cells for relative luciferase activity detection.
MTT assay
After 24 h transfection, these cells were seeded in 96-well plates and cultured for another 24 h. 20 μl (50 mg/ml) of MTT reagent (Sigma-Aldrich, Saint Louis, MO, USA) was added to each complex well at day 1, day 2, day 3, day 4 and day 5. After 4 h incubation and extraction, 150 μl dimethyl sulfoxide (DMSO) reagent (Sigma-Aldrich) was added. We shook the plate for 10 min to fully dissolve the crystallization. The optical absorbance value of each cell group was detected by the enzyme-link meter at 490 nm.
Colony formation assay
After trypsin digestion in 1.5 ml of DMEM containing 10% FBS, 5 × 103 cells in total were suspended. The cells were then cultured in a constant temperature incubator at 5% CO2, 37°C for 14 days. After that, the cells were fixed with paraformaldehyde and stained using 0.1% crystal violet (Sigma-Aldrich). The number of clones was counted under a Nikon Eclipse TS100 microscope (Nikon, Japan).
Wound healing assay
The cells were seeded in a 6-well plate with 2 × 105 cells in each to grow to 80% confluence. We scratched the wound with a pipette tip gently and slowly across the center of the well. After washing three times with serum free medium, cells were incubated in humid atmosphere, 37°C and 5% CO2. The migrated cells were observed with the optical microscope (Nikon, Japan) at 0 h and 24 h respectively.
Transwell assay
30 μl of diluted Matrigel (BD Biosciences, MA, USA) was added into the upper chamber of the transwell and incubated for 30 min at 37°C. After 24 h transfection, cells were digested and added to the upper chamber (1 × 105). 800 μl of medium (containing 5 mg/l fibronectin and calf serum) was subsequently added to the lower chamber for 24 h incubation. Then the cell membrane surface was wiped off with a cotton swab, and the cells were stained with 0.1% crystal violet. The average values of the cells were observed with the optical microscope (Nikon, Japan).
Results
miR-490-3p expression was down-regulated and AURKA expression was up-regulated in hepatocellular carcinoma cells and tissues
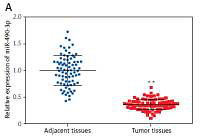
We conducted qRT-PCR to investigate relative expression of miR-490-3p and AURKA, which demonstrated that in HCC tissues, the expression of miR-490-3p was significantly lower and AURKA was remarkably higher, exactly opposite to expression in adjacent tissues. The results of qRT-PCR suggested that miR-490-3p/AURKA might be related to tumor formation (Figures 1 A, B). Western blot results indicated that AURKA protein expression was up-regulated in HCC tissues in comparison to adjacent tissues (Figure 1 C).
Figure 1
Expression of miR-490-3p and aurora kinase A gene (AURKA) in hepatocellular carcinoma (HCC) tissues and cells. A – Relative expression of miR-490-3p in HCC tissues and corresponding adjacent tissues. B – Relative expression of AURKA in HCC tissues and corresponding adjacent tissues. C – Relative expression of AURKA protein in HCC tissues and corresponding adjacent tissues. D – Relative expression of miR-490-3p in HCC cells and normal liver cells. E – Relative expression of AURKA in HCC cells and normal liver cells. F – Relative expression of AURKA protein in HCC cells and normal liver cells
*p < 0.05, **p < 0.01.
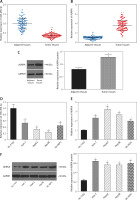
Similarly, qRT-PCR assay and western blot were performed to further detect the relative expression of miR-490-3p and AURKA in 4 kinds of HCC cells (Huh-7, HepG2, Hep3B, SK-HEP-1) and normal hepatic cells (HL-720). The results of both qRT-PCR (Figures 1 D, E) and western blot (Figure 1 F) were identical to the foregoing experiments, proving that miR-490-3p was low-expressed and AURKA was high-expressed in HCC tissues and cells.
In order to explore the relationship between miR-490-3p/AURKA and HCC, we investigated the correlation between miR-490-3p/AURKA expression and clinicopathologic characteristics. The statistical results showed that tumor size, tumor amount, tumor grade and tumor stage were closely associated with expression of miR-490-3p and AURKA (Table II).
Table II
Correlation between miR-490-3p and aurora kinase A gene (AURKA) expression and clinicopathologic characteristics
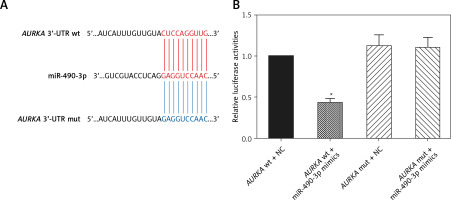
AURKA was the target gene of miR-490-3p
A binding site was found between miR-490-3p and wild type AURKA instead of between miR-490-3p and mutant AURKA (Figure 2 A). To investigate the regulatory relationship between miR-490-3p and AURKA, we transfected miR-490-3p mimics to study the expression of AURKA after transfecting miR-490-3p. It could be seen that the relative luciferase activity of AURKA wt + miR-490-3p mimics significantly decreased compared with the AURKA wt + negative control (NC), AURKA mut + miR-490-3p mimics and AURKA mut + NC, indicating that miR-490-3p could target AURKA and inhibit its expression (Figure 2 B).
Figure 2
Aurora kinase A gene (AURKA) was the target gene of miR-490-3p. A – Binding site of miR-490-3p and AURKA predicted by TargetScan. B – Relative luciferase activity of AURKA wt + miR-490-3p mimic group was significantly lower than AURKA wt + negative control (NC), AURKA mut + NC and AURKA mut + miR-490-3p mimic group
*p < 0.05.
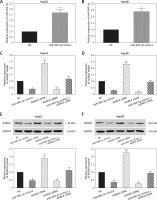
miR-490-3p inhibited expression of AURKA in hepatocellular carcinoma cells
To further detect the relationship between miR-490-3p and AURKA, we transfected miR-490-3p mimics, AURKA siRNA and AURKA cDNA into HepG2 and Hep3B cells. It could be seen that the expression of miR-490-3p was significantly increased after miR-490-4p mimics transfection in both HepG2 and Hep3B cells, showing that the transfection was effective (Figures 3 A, B). qRT-PCR showed that the expression of AURKA was significantly decreased after miR-490-3p mimics or AURKA siRNA expression, while the expression of AURKA was not affected when co-transfected with miR-490-3p mimics and AURKA cDNA (Figures 3 C, D). Western blot showed a parallel result (Figures 3 E, F). All above results demonstrated that AURKA was the target gene of miR-490-3p and overexpression of miR-490-3p could inhibit the expression of AURKA in HCC cells.
Figure 3
miR-490-3p suppresses expression of aurora kinase A gene (AURKA) in hepatocellular carcinoma (HCC) cells. A – qRT-PCR showed that miR-390-3p mimic transfection could significantly increase expression of miR-390-3p in HepG2 cells. B – qRT-PCR showed that miR-390-3p mimic transfection could significantly increase expression of miR- 390-3p in HepG2 cells. C – qRT-PCR showed that AURKA mRNA expression of miR-490-3p mimics and AURKA siRNA group was significantly lower, while AURKA mRNA expression of AURKA cDNA group was higher, and miR-490-3p + AURKA cDNA had no influence on AURKA mRNA expression in HepG2 cells. D – qRT-PCR showed that AURKA mRNA expression of miR-490-3p mimics and AURKA siRNA group was significantly lower, while AURKA mRNA expression of AURKA cDNA group was higher, and miR-490-3p + AURKA cDNA had no influence on the AURKA mRNA expression in Hep3B cells. E – Western blot showed that AURKA protein expression of miR-490-3p mimics and AURKA siRNA group was significantly lower, while AURKA protein expression of AURKA cDNA group was higher, and miR-490-3p + AURKA cDNA had no influence on AURKA protein expression in HepG2 cells. F – Western blot showed that AURKA protein expression of miR-490-3p mimics and AURKA siRNA group was significantly lower, while AURKA protein expression of AURKA cDNA group was higher, and miR-490-3p + AURKA cDNA had no influence on AURKA protein expression in Hep3B cells
*p < 0.05, **p < 0.01, compared with negative control (NC) group.
miR-490-3p/AURKA influence proliferation of HepG2 and Hep3B cells
Colony formation and MTT assay were performed to explore the effect of miR-490-3p/AURKA on proliferation of HCC cells. In the colony formation assay, fewer colonies were detected in the miR-490-3p mimics/AURKA siRNA group, and more colonies were detected in the AURKA cDNA group while the number of cell colonies formed was not changed significantly in the miR-490-3p + AURKA cDNA group (Figures 4 A–C), which suggested that up-regulation of miR-490-3p or down-regulation of AURKA could decrease the proliferation ability of HepG2 and Hep3B cells. Similar to the colony formation assay, the MTT assay result indicated that miR-490-3p mimics and AURKA siRNA could decrease cell viability, while AURKA cDNA would decrease cell viability, and miR-490-3p + AURKA cDNA had no influence on the proliferation of the cells (Figures 4 D, E). Therefore, miR-490-3p could efficiently suppress the proliferation of HepG2 and Hep3B cells via regulating AURKA.
Figure 4
Impact of miR-490-3p and aurora kinase A gene (AURKA) on proliferation of hepatocellular carcinoma (HCC) cells. A – Colony formation images of HepG2 and Hep3B cells showed that colony number of miR-490-3p mimics and AURKA siRNA group was significantly smaller, while colony number area of AURKA cDNA group was larger, and miR-490-3p + AURKA cDNA had no influence on proliferation of the cells. B – Statistical graph of colony formation of HepG2 cells. C – Statistical graph of colony formation of Hep3B cells. D – MTT assay showed that the cell viability of miR-490-3p mimics and AURKA siRNA group was significantly lower, while the cell viability area of AURKA cDNA group was higher, and miR-490-3p + AURKA cDNA had no influence on HepG2 cell viability. E – MTT assay showed that cell viability of miR-490-3p mimics and AURKA siRNA group was significantly lower, while cell viability area of AURKA cDNA group was higher, and miR-490-3p + AURKA cDNA had no influence on Hep3B cell viability
*p < 0.05, **p < 0.01, compared with negative control (NC) group.
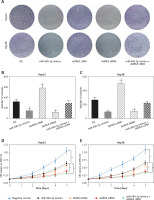
miR-490-3p/AURKA influence migration and invasion of HepG2 and Hep3B cells
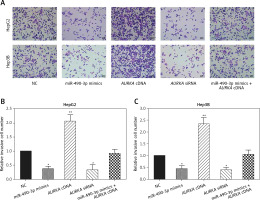
Wound healing assay and transwell assay were performed to detect the migration and invasion ability of HCC cells with miR-490-3p/AURKA regulated. According to the wound healing assay, the closed wounding area in the miR-490-3p mimic group and the AURKA siRNA group was considerably smaller than that in the NC group and displayed a slower speed. In contrast, the AURKA cDNA group presented a quicker wound healing speed, and the effect of AURKA cDNA was counteracted by miR-490-3p (Figures 5 A–C). According to the transwell assay, the number of invasive cells in the miR-490-3p mimic group and AURKA siRNA group was significantly fewer than in the NC group while the number of invasive cells in the AURKA cDNA group drastically increased, and the effect of AURKA cDNA was counteracted by miR-490-3p (Figures 6 A–C). All the above results demonstrated that miR-490-3p overexpression could inhibit migration and invasion of HCC cells, while AURKA reversed the process.
Figure 5
Impact of miR-490-3p and aurora kinase A gene (AURKA) on migration of hepatocellular carcinoma cells. A – Wound healing images of HepG2 and Hep3B cells showed that closed wound area of miR-490-3p mimics and AURKA siRNA group was significantly smaller, while closed wound area of AURKA cDNA group was larger, and miR-490-3p + AURKA cDNA had no influence on migration of the cells. B – Statistical graph of closed wound area of HepG2 cells. C – Statistical graph of closed wound area of Hep3B cells
*p < 0.05, **p < 0.01, compared with negative control (NC) group.
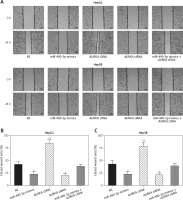
Figure 6
Impact of miR-490-3p and aurora kinase A gene (AURKA) on invasion of hepatocellular carcinoma cells. A – Transwell images of HepG2 and Hep3B cells showed that invasive cell number of miR-490-3p mimics and AURKA siRNA group was significantly decreased, while invasive cell number of AURKA cDNA group was increased, and miR-490-3p + AURKA cDNA had no influence on invasion of the cells. B – Statistical graph of invasive cell number of HepG2 cells. C – Statistical graph of invasive cell number of Hep3B cells
*p < 0.05, **p < 0.01, compared with negative control (NC) group.
Discussion
MiRNA can function as a regulator of tumors by promoting or inhibiting the expression of corresponding tumor-related genes at the post-transcriptional level. It is well established that the aberrant expression of miRNAs is associated with the initiation, progression and prognosis of HCC [23–25]. MiRNA is often reported as a tumor suppressor. Yan et al. reported that miR-140-5p could inhibit HCC by directly targeting the unique isomerase Pin1 and blocking multiple cancer-driving pathways [26]. On the other hand, some studies argued that the overexpression of miRNAs HCC cells and tissues would promote progression of HCC. For instance, Fang et al. found miR-425-5p would accelerate the metastasis and invasion in HCC cells by dysregulating multiple signaling pathways mediated by SCAI [27]. Our study focused on the relationship between the widely studied miRNA miR-490-3p and its target gene, and then discussed how miR-490-3p regulates HCC cell viability.
miR-490-3p is a kind of miRNA which is associated with tumorigenesis and progression of many tumors. These previous studies emphasized miR-490-3p as a tumor suppressor, showing the great research potential of the relationship between miR-490-3p and its target genes. To further investigate the function of miR-490-3p in HCC, we detected the down-regulated miR-490-3p expression in HCC cells and tissues. Furthermore, the overexpression of miR-490-3p could suppress the proliferation, migration and invasion of HCC cells.
Because miRNAs usually carry out their functions via blocking their target genes, we aimed to research the target genes of miR-490 and their interaction. In our study, we confirmed the targeting relationship between miR-490-3p and AURKA via the dual-luciferase reporter assay; we used RNA interference to knock down the expression of AURKA, and miR-490-3p mimics were transfected for overexpression miR-490-3p. The results of molecular experiments (qRT-PCR and western blot) showed that the effect of up-regulating miR-490-3p was the same as silencing AURKA, which immediately verified the result of the dual-luciferase reporter assay. It was confirmed that miR-490-3p could bind to the 3′UTR of AURKA and inhibit its expression, and miR-490-3p could suppress proliferation, migration and invasion of HCC cells through blocking AURKA. However, further exploration about how down-regulated AURKA affects the progress of HCC via regulating its downstream genes or related signaling pathways remains to be performed. Since many genes and signaling pathways affecting the metastasis of tumor cells have been detected and studied [28], the possible relationship among miR-490-3p, AURKA and these downstream genes will be a valuable research topic. In addition, the effect of miR-490-3p and AURKA on the HCC pathologic condition of experimental animals can be explored to further consolidate the conclusion of this study.
In conclusion, miR-490-3p was aberrantly down-regulated while its target gene AURKA was up- regulated in HCC cells. MiR-490-3p served to inhibit HCC proliferation, migration and invasion via blocking AURKA. These findings enhanced our understanding of HCC and provided an in-depth insight into potential HCC treatments. However, the further molecular mechanism of AURKA regulation, the other possible effects of the regulation of miR-490-3p/AURKA, such as apoptosis and autophagy, and further in vivo experiments were not investigated in our work due to the experiment limitations. Therefore, we will consistently focus on the further molecular mechanism of the regulation of miR-490-3p/AURKA and its reflection in living bodies. We hope that it will enrich the theoretical basis of clinical therapy in HCC treatment.