Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
ONCOLOGY / RESEARCH PAPER
Plasma Proteome-Genome Integration Reveals Novel Protein Biomarkers and Drug Targets Linked to Breast Cancer Survival
1
Department of General Surgery, The Second Xiangya Hospital, Central South University, China
2
2. Department of Pharmacy, The Second Xiangya Hospital, Central South University, China
3
Institute of Clinical Pharmacy, Central South University, China
These authors had equal contribution to this work
Submission date: 2025-04-14
Final revision date: 2025-07-06
Acceptance date: 2025-07-14
Online publication date: 2025-08-04
Corresponding author
Shenglan Tan
2. Department of Pharmacy, The Second Xiangya Hospital, Central South University, 410011, Changsha, China
2. Department of Pharmacy, The Second Xiangya Hospital, Central South University, 410011, Changsha, China
KEYWORDS
TOPICS
ABSTRACT
Introduction:
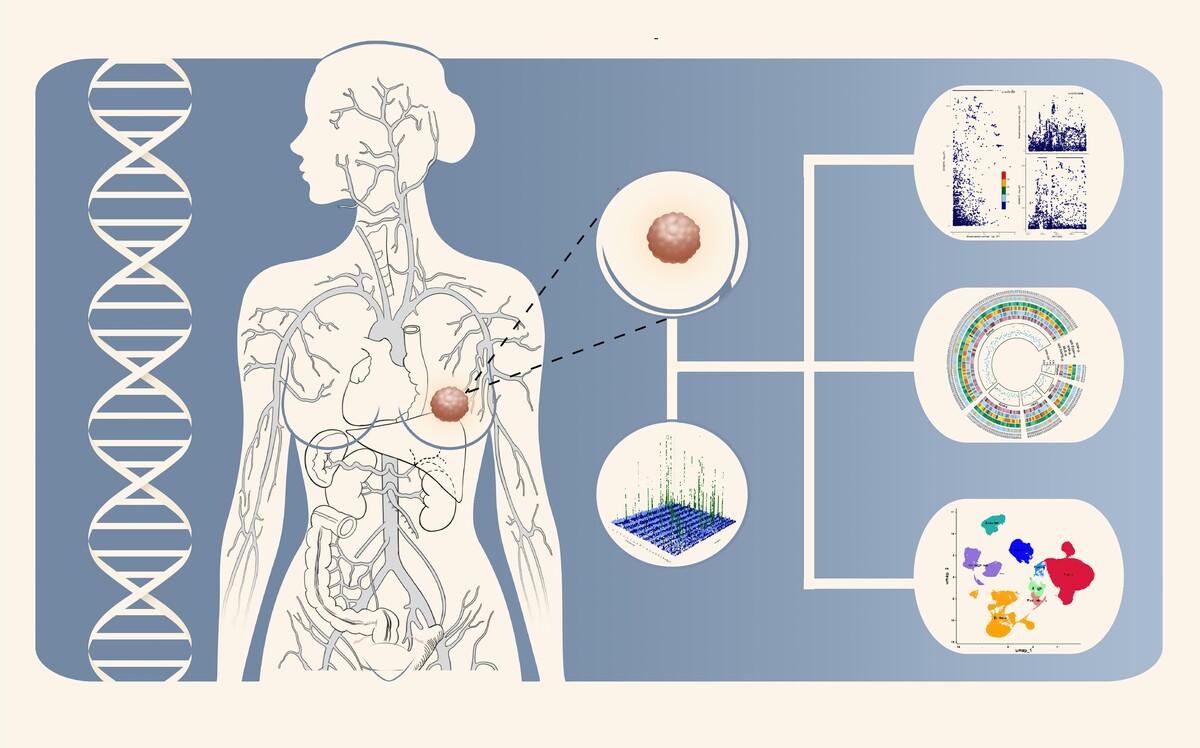
Identifying new drug targets is essential for improving breast cancer survival. The proteome provides a rich source for potential therapeutic targets. This study aimed to identify protein markers and therapeutic targets for breast cancer by using proteome-wide Mendelian randomization (MR).
Material and methods:
Protein quantitative trait loci (pQTL) data were obtained from four large-scaled proteomic studies, including 17,267 circulating protein markers. Genetic associations with breast cancer survival were derived from a large-scale GWAS meta-analysis (37,954 cases, 2,900 deaths). Proteome-wide MR was performed to assess the association between proteins and breast cancer survival, complemented by single-cell expression analysis to identify enriched cell types. Protein-protein interactions (PPI) and druggability assessments were also conducted to prioritize therapeutic targets.
Results:
Gene prediction levels for 27 proteins were found to be associated with breast cancer survival. Among these, eight proteins (ADAM15, CD83, SH3BGRL3, SNCG, ANXA1, GRHPR, ALDH2, and MTHFD2) showed the strongest evidence of association, while four proteins (ARG2, RPL14, NFU1, and TXNL4B) demonstrated a strong but slightly weaker correlation. Notably, SH3BGRL3, GRHPR, ARG2, RPL14, NFU1, and TXNL4B were newly identified as circulating protein markers significantly associated with breast cancer prognosis. Druggability revealed that 13 of these proteins were already targeted by existing drugs, offering potential for breast cancer treatment.
Conclusions:
We identified 27 genes associated with overall and subtype-specific breast cancer survival, providing potential prognostic biomarkers and therapeutic targets, and offering new avenues for improving breast cancer management.
Identifying new drug targets is essential for improving breast cancer survival. The proteome provides a rich source for potential therapeutic targets. This study aimed to identify protein markers and therapeutic targets for breast cancer by using proteome-wide Mendelian randomization (MR).
Material and methods:
Protein quantitative trait loci (pQTL) data were obtained from four large-scaled proteomic studies, including 17,267 circulating protein markers. Genetic associations with breast cancer survival were derived from a large-scale GWAS meta-analysis (37,954 cases, 2,900 deaths). Proteome-wide MR was performed to assess the association between proteins and breast cancer survival, complemented by single-cell expression analysis to identify enriched cell types. Protein-protein interactions (PPI) and druggability assessments were also conducted to prioritize therapeutic targets.
Results:
Gene prediction levels for 27 proteins were found to be associated with breast cancer survival. Among these, eight proteins (ADAM15, CD83, SH3BGRL3, SNCG, ANXA1, GRHPR, ALDH2, and MTHFD2) showed the strongest evidence of association, while four proteins (ARG2, RPL14, NFU1, and TXNL4B) demonstrated a strong but slightly weaker correlation. Notably, SH3BGRL3, GRHPR, ARG2, RPL14, NFU1, and TXNL4B were newly identified as circulating protein markers significantly associated with breast cancer prognosis. Druggability revealed that 13 of these proteins were already targeted by existing drugs, offering potential for breast cancer treatment.
Conclusions:
We identified 27 genes associated with overall and subtype-specific breast cancer survival, providing potential prognostic biomarkers and therapeutic targets, and offering new avenues for improving breast cancer management.
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.