Introduction
Breast cancer occurs most among females, and is the main cause of cancer-related mortality in women all over the world [1]. The development of breast cancer is very complicated, and includes environmental and genetic elements. As one of the genetic variants, single nucleotide polymorphisms (SNPs) are widely used to predict the disease risk, prognosis, and clinical outcome [2, 3]. Accumulative investigations have found that SNPs in tumor oncogenes or suppressor genes could play a vital role in breast cancer genetic susceptibility [4–13]. Long non-coding RNAs (lncRNAs) are RNA molecules which exceed 200 nucleotides (nt), but they are unable to encode protein [14]. Growing evidence suggests that LncRNAs interact with DNA, RNA and protein so that they can regulate gene expression at transcriptional and post-transcriptional levels [15, 16].
HOX transcript antisense intergenic RNA (HOTAIR) is a non-coding RNA transcribed from the HOXC locus with an approximate length of 2.2 kb. The long arm of chromosome 12 (12q13.13) is where the HOTAIR gene is located. As an oncogene, HOTAIR is crucial in gene and chromatin regulation, and it is persistently overexpressed in various cancers, involved in tumor invasion, and associated with bad prognosis of corresponding cancers [2, 3, 17–22].
Recently, the relations of HOTAIR SNPs with breast tumor risk have been investigated [19, 23–25]. However, some conclusions remain controversial. Obviously, it is still necessary to further analyze the relation between HOTAIR SNPs and breast cancer susceptibility. Therefore, a meta-analysis of all eligible published case-control studies was conducted to assess the effect of four lncRNA HOTAIR SNPs (rs1899663, rs4759314, rs920778 and rs12826786) on breast cancer risk.
Material and methods
Search strategy and eligibility criteria
Electronic databases (Embase, PubMed, Cochrane Library databases, and Web of Science) were searched by us until July 16, 2019 using the following key words: “HOTAIR or HOX transcript antisense RNA or LncRNA HOTAIR” and “Polymorphism or variation or mutation or genotypes or SNP” and “breast cancer or breast tumor or breast carcinoma”. In addition, a manual search was needed for references of relevant articles in order to achieve potential eligible publications. Studies were included in our meta-analysis if the following criteria were met: (1) case-control studies concerned with the association between HOTAIR polymorphisms and breast cancer, (2) published data related to the frequencies of alleles or genotypes must be sufficient, (3) all studies were published in English. Exclusion criteria included: (1) meta-analysis, reviews and letters, (2) no more than two studies assessed one LncRNA HOTAIR gene.
Data extraction and quality evaluation
Two investigators (Wang B and Yuan FL) independently extracted the following data from each publication: First author, Year, Racial descent, Country, Source of controls, Quantities of cases and controls, Genotype distributions of cases and controls. Diverse racial descendants were classified as West Asian and East Asian. Disputes were resolved by engaging a third investigator.
Statistical analysis
Crude odds ratios (ORs) with 95% confidence interval (95% CI) were used to calculate and evaluate the strength of relations between lncRNA HOTAIR polymorphisms and breast cancer susceptibility. The pooled ORs were estimated according to five different comparison models (allele contrast, dominant, recessive, homozygous and heterozygous). The heterogeneity between each study was assessed with the χ2-based Q statistic test. If the p-value was less than 0.1, significant heterogeneity was found and the random effect (DerSimonian-Laird method) model was applied; otherwise, a fixed effect (Mantel-Haenszel method) model was used. In this meta-analysis, we went a step further to examine the ethnicities, source of controls and genotyping methods so that we could explore the source of heterogeneity. Sensitivity analysis was conducted through removing each study in turn to evaluate the stability of our results. Publication bias was evaluated by funnel plot and Egger’s test. All data analyses were carried out using the Stata 12.0 software. P-values < 0.05 were considered statistically significant.
Results
Study characteristics
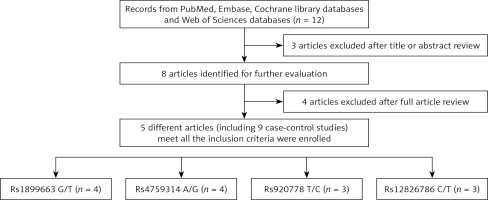
Through searching the Embase, PubMed, Cochrane Library databases, and Web of Science using the key words, we obtained 12 articles which focused on the association between the LncRNA HOTAIR polymorphisms and susceptibility to breast cancer. According to Figure 1, in all, 4 studies with 4936 cases and 5214 controls conformed to the inclusion criteria and four HOTAIR SNPs were included in this meta-analysis [23–26]. The chief characteristics of three HOTAIR SNPs are shown in Table I. 1813 cases and 1904 controls were contained in 4 studies for rs1899663 and rs4759314 polymorphisms. As for the rs920778 polymorphism, there were 845 cases and 856 controls. As for rs12826786, there were 465 cases and 550 controls. Racial descent came from Asia. The main countries were Iran, China and Turkey. According to the source of control, all studies were defined as hospital-based or population-based. Genotype methods included PCR-RFLP, PCR-sequencer, TaqMan, and CRS-RFLP. Four HOTAIR SNPs were extracted from all eligible studies. Genotype distributions of HOTAIR rs1899663, rs4759314, rs920778, rs12826786 are listed in Table II. Each has one study in three SNPs (rs1899663, rs4759314, rs920778) deviating from Hardy-Weinberg equilibrium (HWE).
Table I
Main characteristics of all included studies
Table II
Characteristics of case-control studies included in the meta-analysis
Quantitative synthesis
The main meta-analysis results are shown in Table III. Five different comparison models were used to evaluate the pooled ORs. No significance was found in all genetic models of all three SNPs. To go a step further, the data were stratified into different subgroups in the light of ethnicity, source of controls and genotyping methods. There was no significance in the subgroup analysis of rs4759314 polymorphism either (Figure 2, Table III).
Table III
Summary ORs and 95% CI of HOTAIR polymorphisms and breast cancer risk
Figure 2
Forest plot of the association between HOTAIR polymorphisms and breast cancer risk. A – rs1899663 in allele model; B – rs4759314 in allele model C – rs920778 in recessive model; D – rs12826786 in heterozygous model

As for the rs1899663 polymorphism, there was no significance in the ethnicity subgroups. There was only one study about a hospital-based control subgroup. Increasing risks were found in the allele model, recessive model, homozygous model and heterozygous model with a hospital-based control subgroup (T vs. C: 1.22, 95% CI: 1.03–1.44, p = 0.02; TT vs. CC + CT: 2.00, 95% CI: 1.19–3.62, p = 0.008; TT vs. CC: 2.12, 95% CI: 1.24–3.42, p = 0.006; TT vs. CT: 1.9, 95% CI: 1.08–2.95, p = 0.023) (Figure 2, Table III).
As for the rs920778 polymorphism, it was interesting to find that decreasing risks were observed in the West Asian subgroup under recessive, homozygous and heterozygous models (TT vs. CC + CT: 3.33, 95% CI: 1.65–6.72, p = 0.001; TT vs. CC: 3.96, 95% CI: 1.13–13.85, p = 0.03; TT vs. CT: 2.94, 95% CI: 1.71–5.03, p < 0.0001). Meanwhile, increasing risks were found in the East Asian subgroup under allele and dominant models (T vs. C: 0.78, 95% CI: 0.65–0.93, p = 0.005; CT + TT vs. CC: 0.68, 95% CI: 0.53–1.89, p = 0.004). Additionally, the associations between HOTAIR and the breast cancer risk were found to be significant in the hospital-based control subgroup (TT vs. CC + CT: 2.4, 95% CI: 1.22–4.73, p = 0.01; TT vs. CT: 2.62, 95% CI: 1.28–5.37, p = 0.008) (Figure 2, Table III).
As for the rs12826786 polymorphism, racial descent was all from west Asia. Similarly to the rs1899663 polymorphism, there was also one study about a hospital-based control subgroup. Increasing risks were found in the recessive model, homozygous model and heterozygous model with the hospital-based control subgroup (TT vs. CC + CT: 2.49, 95% CI: 1.25–4.97, p = 0.01; TT vs. CC: 2.25, 95% CI: 1.05–4.81, p = 0.038; TT vs. CT: 2.69, 95% CI: 1.29–5.56, p = 0.008) (Figure 2, Table III).
Publication bias and sensitivity analysis
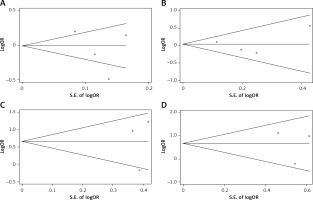
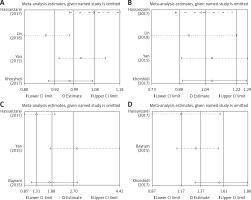
Egger’s test and Begg’s funnel plot were used so that we could evaluate the potential publication bias of the studied literature. No obvious evidence of publication bias was detected in interleukin-1β (rs1899663, rs4759314, rs920778 and rs12826786) (Figure 3). Also, it showed good results in the sensitivity analysis (Figure 4).
Discussion
HOTAIR expression has been considered to be a new prognostic biomarker for primary breast cancer. GEPIA database (http://gepia.cancer-pku.cn/index.html) analysis based on data from The Cancer Genome Atlas (TCGA) showed that HOTAIR expression only existed in the mammary gland of BR patients. Meanwhile, the HOTAIR mRNA levels in the BR patients were significantly highly than in the controls. Subsequently, the circlncRNAnet database CirclncRNAnet (http://app.cgu.edu.tw/circlnc/) showed that lncRNA HOTAIR in BR patients was 4 times higher than in the normal patients and interacted with numerous miRNAs.
Emerging studies have concentrated on links between HOTAIR polymorphisms (rs1899663, rs4759314, rs920778 and rs12826786) and cancer risk. Meanwhile, several meta-analyses investigated the relations of HOTAIR polymorphisms and cancer risk [27–33]; however, none of them have focused on the breast cancer risk independently due to lack of eligible data and some results remain confusing. For example, Qi et al. reported that the HOTAIR rs920778 polymorphism increased cancer risk under allele and recessive models in overall analyses while Tian et al. failed to observe significant associations between them [29, 33]. Zhang et al. reported that the rs920778 polymorphism was relevant to cancer susceptibility in Asian population but not in Turks [30]. Chu et al. summarized the results indicating that the rs920778 polymorphism could increase cancer risks under a recessive model [31]. Liu et al. detected the rs920778 polymorphism, finding that it was closely related to increased cancer risk in all genetic models, but no significant association between rs1899663 polymorphism and cancer risk was observed [32]. Li’s meta-analysis revealed a significant association between HOTAIR rs4759314 and susceptibility to breast cancer; however, we failed to corroborate that in our meta-analysis [34]. Therefore, a meta-analysis was needed.
To our knowledge, it is for the first time that such a meta-analysis has been used to evaluate the linkages between LncRNA HOTAIR polymorphisms and breast cancer susceptibility in overall and stratified analyses. Generally, our results showed no major relations of HOTAIR (rs1899663, rs4759314, rs920778 and rs12826786) polymorphisms and breast cancer risk. In terms of rs4759314 polymorphism, similar results with no significant association were found in subgroup analyses. As for rs1899663 polymorphism, there was decreasing breast cancer risk in the hospital-based control subgroup. As for the rs920778 polymorphism, in terms of ethnicity subgroup analysis, it was interesting to find that the results of West Asia and East Asia were contrary. As for rs12826786, it decreased breast cancer risk in the hospital-based control subgroup.
Although we found that LncRNA HOTAIR polymorphisms were closely related to breast cancer risk in our meta-analysis, there are some limitations to our study that should be taken into consideration. First, the sample size is relatively small in subgroup analysis. Second, only Asians were included in our meta-analysis, which will produce publication bias. Third, in one study on each of three SNPs (rs1899663, rs4759314, rs920778) of HOTAIR the data deviate from HWE. Finally, further analysis was limited by lack of detailed information and original data.
In conclusion, our current meta-analysis found significant links of HOTAIR polymorphisms and the breast cancer risk among Asian people, suggesting that HOTAIR rs920778, rs1899663 and rs12826786 polymorphisms may contribute to breast cancer susceptibility. In future, more comprehensive studies and a large number of samples are needed to verify this association.