Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
ONCOLOGY / CLINICAL RESEARCH
miRNA-mediated regulation of extracellular matrix dynamics across breast cancer subtypes
1
Department of Plastic Surgery, Faculty of Medicine, Academia of Silesia, Katowice, Poland
2
Department of Plastic and Reconstructive Surgery, Hospital for Minimally Invasive and Reconstructive Surgery in Bielsko-Biala, Bielsko-Biala, Poland
3
Department of Medical and Health Sciences, Collegium Medicum, WSB University, Dabrowa Gornicza, Poland
4
Independent Researcher, Wloclawek, Poland
5
Faculty of Medicine and Health Sciences, Andrzej Frycz Modrzewski University in Krakow, Krakow, Poland
6
Department of Gynecology and Obstetrics, TOMMED Specjalisci od Zdrowia, Katowice, Poland
7
Chalcarz Clinic-Aesthetic Surgery, Aesthetic Medicine, Poznan, Poland
8
Bieńkowski Medical Center-Plastic Surgery, Bydgoszcz, Poland
9
New Medical Techniques Specjalist Hospital of St. Family in Rudna Mała, Rzeszow, Poland
10
Department of Histology and Cell Pathology, Faculty of Medical Sciences in Zabrze, Medical University of Silesia in Katowice, Poland
Submission date: 2025-02-06
Final revision date: 2025-05-18
Acceptance date: 2025-05-30
Online publication date: 2025-07-10
Corresponding author
Tomasz Sirek
Department of Plastic Surgery, Faculty of Medicine, Academy of Silesia, 40-555, Katowice, Poland
Department of Plastic Surgery, Faculty of Medicine, Academy of Silesia, 40-555, Katowice, Poland
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Interactions between extracellular signals and the extracellular matrix (ECM) influence cellular phenotype and molecular functions, affecting proliferation, differentiation, adhesion, apoptosis, and migration. Deregulation of ECM remodeling contributes to the development of diseases, including breast cancer. The study was aimed to identify microRNAs (miRNAs) that may regulate the activity of genes involved in ECM remodeling and focal adhesion across five breast cancer subtypes in Polish women.
Material and methods:
The study enrolled patients representing five breast cancer subtypes: 130 luminal A, 100 HER2-negative luminal B, 96 HER2-positive luminal B, 36 non-luminal HER2-positive, 43 triple-negative breast cancer (TNBC) cases. Cancer tissue samples were collected during surgery along with healthy tissue margins (control group). The expression profiles of genes associated with ECM remodeling and focal adhesion were evaluated with mRNA microarrays and reverse transcription quantitative polymerase chain reaction (RT-qPCR). Protein expression was assessed using enzyme-linked immunosorbent assay (ELISA). miRNA detection and target prediction were performed using miRNA microarrays.
Results:
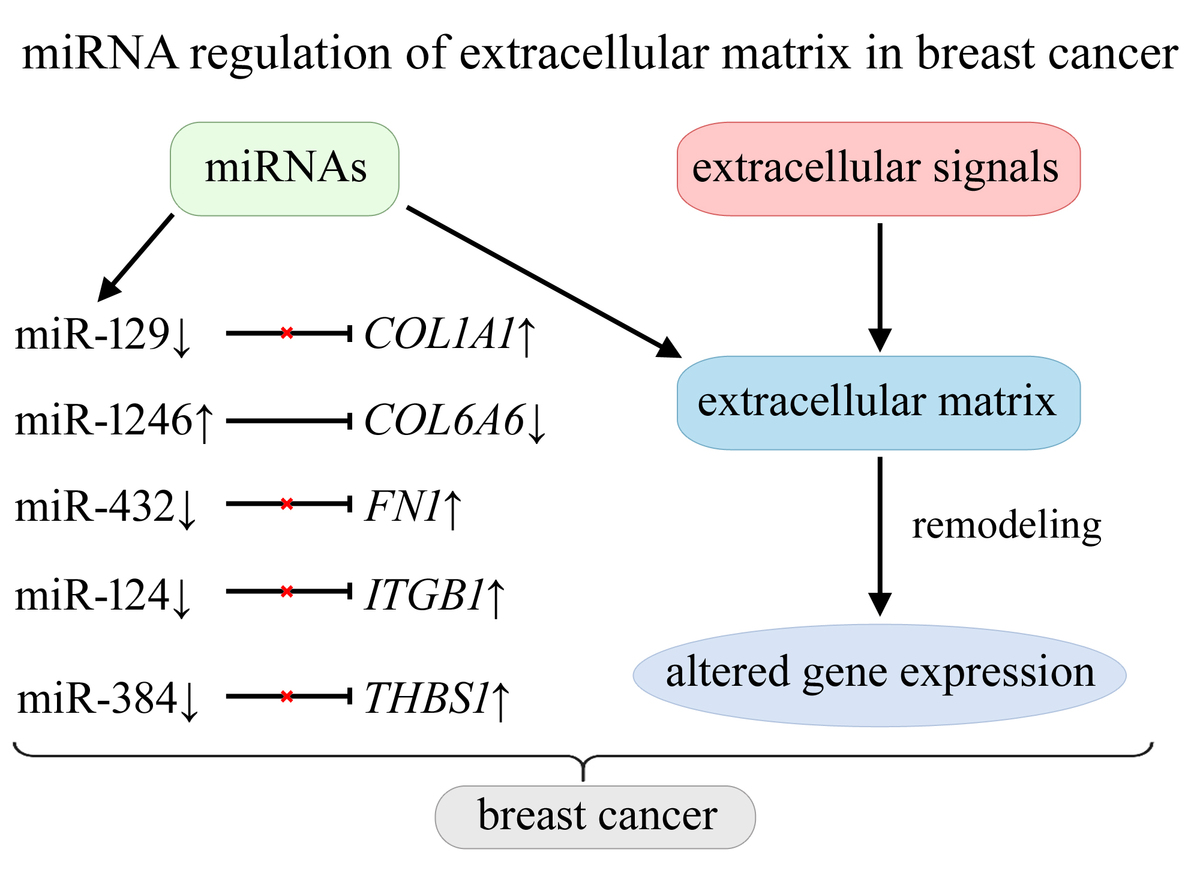
Overexpression of COL1A1, FN1, ITGB1, and THBS1 may be associated with reduced levels of miR-129, miR-432, miR-124, and miR-384, respectively. Decreased COL6A6 expression may result from increased activity of miR-1246. Additionally, the study revealed increased levels of COL1A2, COMP, and SPP1 with reduced activity of RELN, across all five breast cancer subtypes.
Conclusions:
This is the first study to comprehensively analyze miRNA-mediated regulation of ECM-related genes across five breast cancer subtypes in a Polish cohort. Overexpression of COL1A1, FN1, and ITGB1 is linked to reduced levels of specific miRNAs, while decreased COL6A6 expression is associated with increased miR-1246 activity.
Interactions between extracellular signals and the extracellular matrix (ECM) influence cellular phenotype and molecular functions, affecting proliferation, differentiation, adhesion, apoptosis, and migration. Deregulation of ECM remodeling contributes to the development of diseases, including breast cancer. The study was aimed to identify microRNAs (miRNAs) that may regulate the activity of genes involved in ECM remodeling and focal adhesion across five breast cancer subtypes in Polish women.
Material and methods:
The study enrolled patients representing five breast cancer subtypes: 130 luminal A, 100 HER2-negative luminal B, 96 HER2-positive luminal B, 36 non-luminal HER2-positive, 43 triple-negative breast cancer (TNBC) cases. Cancer tissue samples were collected during surgery along with healthy tissue margins (control group). The expression profiles of genes associated with ECM remodeling and focal adhesion were evaluated with mRNA microarrays and reverse transcription quantitative polymerase chain reaction (RT-qPCR). Protein expression was assessed using enzyme-linked immunosorbent assay (ELISA). miRNA detection and target prediction were performed using miRNA microarrays.
Results:
Overexpression of COL1A1, FN1, ITGB1, and THBS1 may be associated with reduced levels of miR-129, miR-432, miR-124, and miR-384, respectively. Decreased COL6A6 expression may result from increased activity of miR-1246. Additionally, the study revealed increased levels of COL1A2, COMP, and SPP1 with reduced activity of RELN, across all five breast cancer subtypes.
Conclusions:
This is the first study to comprehensively analyze miRNA-mediated regulation of ECM-related genes across five breast cancer subtypes in a Polish cohort. Overexpression of COL1A1, FN1, and ITGB1 is linked to reduced levels of specific miRNAs, while decreased COL6A6 expression is associated with increased miR-1246 activity.
REFERENCES (68)
1.
Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024; 74: 229-63.
3.
Yang X, Smirnov A, Buonomo OC, et al. A primary luminal/HER2 negative breast cancer patient with mismatch repair deficiency. Cell Death Discov 2023; 9: 365.
4.
Łukasiewicz S, Czeczelewski M, Forma A, et al. Breast cancer-epidemiology, risk factors, classification, prognostic markers, and current treatment strategies-an updated review. Cancers (Basel) 2021; 13: 4287.
5.
Yang ZJ, Liu YX, Huang Y, et al. The regrouping of Luminal B (HER2 negative), a better discriminator of outcome and recurrence score. Cancer Med 2023; 12: 2493-504.
6.
Falato C, Schettini F, Pascual T, et al. Clinical implications of the intrinsic molecular subtypes in hormone receptor-positive and HER2-negative metastatic breast cancer. Cancer Treat Rev 2023; 112: 102496.
7.
Thomas A, Reis-Filho JS, Geyer CE, et al. Rare subtypes of triple negative breast cancer: current understanding and future directions. NPJ Breast Cancer 2023; 9: 55.
8.
Poltavets V, Kochetkova M, Pitson SM, et al. The role of the extracellular matrix and its molecular and cellular regulators in cancer cell plasticity. Front Oncol 2018; 8: 431.
9.
Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, et al. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun 2020; 11: 5120.
10.
Sainio A, Järveläinen H. Extracellular matrix-cell interactions: focus on therapeutic applications. Cell Signal 2020; 66: 109487.
11.
Mouw JK, Ou G, Weaver VM. Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Biol 2014; 15: 771-85.
12.
Siddhartha R, Garg M. Interplay between extracellular matrix remodeling and angiogenesis in tumor ecosystem. Mol Cancer Ther 2023; 22: 291-305.
13.
Pramanik D, Jolly MK, Bhat R. Matrix adhesion and remodeling diversifies modes of cancer invasion across spatial scales. J Theor Biol 2021; 524: 110733.
14.
Mohan V, Das A, Sagi I. Emerging roles of ECM remodeling processes in cancer. Semin Cancer Biol 2020; 62: 192-200.
15.
Shi R, Zhang Z, Zhu A, et al. Targeting type I collagen for cancer treatment. Int J Cancer 2022; 151: 665-83.
16.
Li X, Sun X, Kan C, et al. COL1A1: a novel oncogenic gene and therapeutic target in malignancies. Pathol Res Pract 2022; 236: 154013.
17.
Jena MK, Janjanam J. Role of extracellular matrix in breast cancer development: a brief update. F1000Res 2018; 7: 274.
18.
Acerbi I, Cassereau L, Dean I, et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (Camb) 2015; 7: 1120-34.
19.
Cao H, Lee MKH, Yang H, et al. Mechanoregulation of Cancer-associated fibroblast phenotype in three-dimensional interpenetrating hydrogel networks. Langmuir 2019; 35: 7487-95.
20.
Zhao Y, Zheng X, Zheng Y, et al. Extracellular matrix: emerging roles and potential therapeutic targets for breast cancer. Front Oncol 2021; 11: 650453.
21.
Ghafouri-Fard S, Khanbabapour Sasi A, Abak A, et al. Contribution of miRNAs in the pathogenesis of breast cancer. Front Oncol 2021; 11: 768949.
22.
Liu LL, Zhao H, Ma TF, et al. Identification of valid reference genes for the normalization of RT-qPCR expression studies in human breast cancer cell lines treated with and without transient transfection. PLoS One 2015; 10: e0117058.
23.
Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020; 48: D127-31.
26.
Győrffy B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation (Camb) 2024; 5: 100625.
27.
Győrffy B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br J Pharmacol 2024; 181: 362–374.
28.
Kong Y, Yang L, Wei W, et al. CircPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics 2019; 11: 1163-76.
29.
Wu S, Lu J, Zhu H, et al. A novel axis of circKIF4A-miR-637-STAT3 promotes brain metastasis in triple-negative breast cancer. Cancer Lett 2024; 581: 216508.
30.
Lu Y, Wang K, Peng Y, et al. Hsa-miR-214-3p inhibits breast cancer cell growth and improves the tumor immune microenvironment by downregulating B7H3. Oncol Res 2025; 33: 103-21.
31.
Liu J, Shen JX, Wu HT, et al. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med 2018; 25: 211-23.
32.
Ma B, Li F, Ma B. Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer. Open Life Sci 2023; 18: 20220776.
33.
Yang Z, Sun R, Qu G, et al. Identification of key genes in HER2-positive breast cancer with brain metastasis via bioinformatics methods. Transl Cancer Res 2023; 12: 1112-27.
34.
Meng R, Fang J, Yu Y, et al. miR-129-5p suppresses breast cancer proliferation by targeting CBX4. Neoplasma 2018; 65: 572-8.
35.
Zeng H, Wang L, Wang J, et al. microRNA-129-5p suppresses Adriamycin resistance in breast cancer by targeting SOX2. Arch Biochem Biophys 2018; 651: 52-60.
36.
Tang Q, Zhou F, Yang C, et al. CircINTS4 facilitates chemoresistance of TNBC by competitively binding miR-129-5p/POM121 Axis. J Oncol 2022; 2022: 2630864.
37.
Li Q, Gu Z, Tan Q, et al. MicroRNA-129-1-3p represses the progression of triple-negative breast cancer by targeting the GRIN2D gene. Biomed Res Int 2022; 2022: 1549357.
38.
Setijono SR, Park M, Kim G, et al. miR-218 and miR-129 regulate breast cancer progression by targeting lamins. Biochem Biophys Res Commun 2018; 496: 826-33.
39.
Long R, Liu Z, Li J, et al. COL6A6 interacted with P4HA3 to suppress the growth and metastasis of pituitary adenoma via blocking PI3K-Akt pathway. Aging (Albany NY) 2019; 11: 8845-59.
40.
Ma Y, Qiu M, Guo H, et al. Comprehensive analysis of the immune and prognostic implication of COL6A6 in lung adenocarcinoma. Front Oncol 2021; 11: 633420.
41.
Srour MK, Gao B, Dadmanesh F, et al. Gene expression comparison between primary triple-negative breast cancer and paired axillary and sentinel lymph node metastasis. Breast J 2020; 26: 904-10.
42.
Yeh MH, Tzeng YJ, Fu TY, et al. Extracellular matrix-receptor interaction signaling genes associated with inferior breast cancer survival. Anticancer Res 2018; 38: 4593-605.
43.
Huynh KQ, Le AT, Phan TT, et al. The diagnostic power of circulating miR-1246 in screening cancer: an updated meta-analysis. Oxid Med Cell Longev 2023; 2023: 8379231.
44.
Dai Y, Pan Y, Quan M, et al. MicroRNA-1246 mediates drug resistance and metastasis in breast cancer by targeting NFE2L3. Front Oncol 2021; 11: 677168.
45.
Wang P, Chen W, Zhang Y, et al. MicroRNA-1246 suppresses the metastasis of breast cancer cells by targeting the DYRK1A/PGRN axis to prevent the epithelial-mesenchymal transition. Mol Biol Rep 2022; 49: 2711-21.
46.
Scognamiglio I, Cocca L, Puoti I, et al. Exosomal microRNAs synergistically trigger stromal fibroblasts in breast cancer. Mol The Nucleic Acids 2022; 28: 17-31.
47.
Hanitrarimalala V, Bednarska I, Murakami T, et al. Intracellular cartilage oligomeric matrix protein augments breast cancer resistance to chemotherapy. Cell Death Dis 2024; 15: 480.
48.
Papadakos KS, Darlix A, Jacot W, et al. High levels of cartilage oligomeric matrix protein in the serum of breast cancer patients can serve as an independent prognostic marker. Front Oncol 2019; 9: 1141.
49.
Papadakos KS, Hagerling C, Rydén L, et al. High levels of expression of cartilage oligomeric matrix protein in lymph node metastases in breast cancer are associated with reduced survival. Cancers (Basel) 2021; 13: 5876.
50.
Li Y, Hua K, Jin J, et al. miR-497 inhibits proliferation and invasion in triple-negative breast cancer cells via YAP1. Oncol Lett 2021; 22: 580.
51.
Göthlin Eremo A, Lagergren K, Othman L, et al. Evaluation of SPP1/osteopontin expression as predictor of recurrence in tamoxifen treated breast cancer. Sci Rep 2020; 10: 1451.
52.
Pan H, Luo Z, Lin F, et al. FN1, a reliable prognostic biomarker for thyroid cancer, is associated with tumor immunity and an unfavorable prognosis. Oncol Lett 2024; 28: 510.
53.
Chen C, Ye L, Yi J, et al. Correction: FN1-mediated activation of aspartate metabolism promotes the progression of triple-negative and luminal a breast cancer. Breast Cancer Res Treat 2024; 204: 425-7.
54.
Zhang XX, Luo JH, Wu LQ. FN1 overexpression is correlated with unfavorable prognosis and immune infiltrates in breast cancer. Front Genet 2022; 13: 913659.
55.
Huang S, Huang P, Wu H, et al. LINC02381 aggravates breast cancer through the miR-1271-5p/FN1 axis to activate PI3K/AKT pathway. Mol Carcinog 2022; 61: 346-58.
56.
Yang X, Hu Q, Hu LX, et al. miR-200b regulates epithelial-mesenchymal transition of chemo-resistant breast cancer cells by targeting FN1. Discov Med 2017; 24: 75-85.
57.
Wu J, Zhou Z. MicroRNA-432 acts as a prognostic biomarker and an inhibitor of cell proliferation, migration, and invasion in breast cancer. Clin Breast Cancer 2021; 21: e462-70.
58.
Su C, Mo J, Dong S, et al. Integrin-1 in disorders and cancers: molecular mechanisms and therapeutic targets. Cell Commun Signal 2024; 22: 71.
59.
Dittmer A, Dittmer J. A CAF-fueled TIMP-1/CD63/ITGB1/STAT3 feedback loop promotes migration and growth of breast cancer cells. Cancers 2022; 14: 4983.
60.
Rana PS, Wang W, Markovic V, et al. The WAVE2/miR-29/integrin-1 oncogenic signaling axis promotes tumor growth and metastasis in triple-negative breast cancer. Cancer Res Commun 2023; 3: 160-74.
61.
Pang Y, Wu J, Li X, et al. NEAT1/miR 124/STAT3 feedback loop promotes breast cancer progression. Int J Oncol 2019; 55: 745-54.
62.
Cha N, Jia B, He Y, et al. MicroRNA-124 suppresses the invasion and proliferation of breast cancer cells by targeting TFAP4. Oncol Lett 2021; 21: 271.
63.
Liu C, Xing H, Guo C, et al. MiR-124 reversed the doxorubicin resistance of breast cancer stem cells through STAT3/HIF-1 signaling pathways. Cell Cycle 2019; 18: 2215-27.
64.
Kaur S, Bronson SM, Pal-Nath D, et al. Functions of thrombospondin-1 in the tumor microenvironment Int J Mol Sci 2021; 22: 4570.
65.
Li Y, Qin J, Chen G, et al. Plasma THBS1 as a predictive biomarker for poor prognosis and brain metastasis in patients with HER2-enriched breast cancer. Int J Clin Oncol 2024; 29: 427-41.
66.
Marcheteau E, Farge T, Pérès M, et al. Thrombospondin-1 silencing improves lymphocyte infiltration in tumors and response to anti-PD-1 in triple-negative breast cancer. Cancers (Basel) 2021; 13: 4059.
67.
Ma Q, Qi X, Lin X, et al. LncRNA SNHG3 promotes cell proliferation and invasion through the miR-384/hepatoma-derived growth factor axis in breast cancer. Hum Cell 2020; 33: 232-42.
68.
Wang Q, Liang D, Shen P, et al. Hsa_circ_0092276 promotes doxorubicin resistance in breast cancer cells by regulating autophagy via miR-348/ATG7 axis. Transl Oncol 2021; 14: 101045.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.