Introduction
Uterine leiomyomas (UL) arise from the overgrowth of smooth muscle and connective tissues in the uterus. Although these tumors are benign in nature, they account for different co-morbidities in 70% to 80% of premenopausal women [1, 2]. Leiomyomas may be single or multiple and are classified into four types according to their location, i.e. intramural, subserosal, submucosal and cervical. Clinical symptoms vary between patients, and the most common presentation includes heavy vaginal bleeding, pelvic pain, compression of the ureter and pregnancy complications. Different potential risk factors including obesity, nulliparity, early menarche, age, ethnicity and genetic factors are proposed to contribute in the development of UL [3].
Genetic studies have identified that approximately 40% to 50% of UL tissue samples carry chromosomal abnormalities. The most common abnormality observed in larger fibroids is the chromosomal translocation (t(12;14) (q14-15; q23-24)) [4, 5]. Abnormalities in other chromosomes such as, 2, 7, 8, 12, and 22 were also reported [6]. Additionally, a spectrum of mutations in several genes, including high mobility group AT-hook 2 (HMGA2), mediator complex subunit 12 (MED12), fumarate hydratase (FH), collagen, type IV, alpha 5 and alpha 6 (COL4A5-COL4A6), MTND1 and MTCYB were also identified [7–11]. HMGA2 and MED12 represent the two most common genes with mutations in 80–90% of all UL [3]. The role of epigenetic factors may also be an underlying cause for fibroid development, as evidenced by the tumor specific hypomethylation DNA (DNMT1 and DNMT3A genes) [12]. Comparative genomic hybridization (CGH) analysis has also shown an inverse association between the expression status of miRNA and mRNA genes involved signaling pathways, which are important for cell division and growth. These pathways include WNT, JAK, MAPK, STAT and TGF-B [13, 14]. Also, a genome-wide association study (GWAS) suggested significant association of UL with 3 SNPs (rs7913069, rs12484776 and rs2280543) [15]. A study from KSA further confirms the contribution of rs12484776 to leiomyoma risk among Saudi women [16].
Leiomyomas grow during the reproductive phase and shrink or disappear after menopause, suggesting that they are hormone responsive tumors. Circulating levels of sex steroid hormones such as estrogen, progesterone and prolactin levels are elevated in leiomyoma patients [17, 18]. The specific molecular causes underlying the elevated hormone levels could be attributed to different molecular anomalies including somatic mutations [19], gene polymorphisms [20–22], epigenetics or miRNA related gene expression changes [23]. Over-expression of the prolactin (PRL) gene in leiomyomas is reported in tumor samples [3]. The role of PRL gene somatic mutations is well characterized in a variety of gynecological cancers, such as endometroid adenocarcinomas and cervical carcinomas [24]. However, data on the role of somatic mutations in the PRL gene in leiomyomas and their role in influencing circulating prolactin levels is not available. Therefore, in order to explore the role of somatic mutations in the PRL gene, this study screened all the coding regions of the prolactin gene in leiomyomas and matched myometrium tissues. Furthermore, we measured the serum prolactin level in leiomyoma patients and assessed its relationship with PRL gene mutation status and phenotypic features.
Material and methods
Clinical recruitment
This study was approved by the Institutional Ethics Committee on Human Research of King Abdulaziz University Hospital (KAUH), Jeddah. This study recruited fifty Saudi patients clinically diagnosed with leiomyomas through ultrasonography and confirmed through histopathological examination. Additionally, fifty Saudi female control participants, who were confirmed to be free from UL through ultrasonography, were also recruited to assess their serum prolactin levels. Exclusion criteria included pregnant and lactating women, carriers of any gynecological and pituitary gland tumors and those undergoing hormonal therapy. They were mostly of premenopausal age. Clinical data (symptoms and age) and tumor phenotypes (size, number and location) were obtained only for the patients, whereas the basic anthropometric details (height, weight and BMI) and serum prolactin levels were obtained for both patients and control participants. These data were collected through the hospital e-medical records.
Sampling
A total of 100 tissue samples (consisting of both leiomyomas and their corresponding myometrium) were obtained from 50 hysterectomized uteri from the Department of Histopathology at KAUH, Jeddah. Tissues (1–15 mg) were collected in small vials filled with normal saline and stored at –70°C until they were processed for genetic analysis. Tissue samples were collected solely from the patients. For prolactin serum level analysis, 5 ml venous blood samples were withdrawn from all participants (50 UL patients and 50 healthy controls); serum was separated and stored at –20°C, until further use.
Genetic analysis
Genomic DNA isolation
Initially, myometrium and leiomyoma tissues were minced and then DNA extraction was carried out using the QIAamp DNA Tissue extraction kit following the manufacturer’s instructions (Qiagen, Alameda, CA, USA). The concentration and purity of DNA were estimated using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Inc.) at an absorbance of 260 nm. Extracted DNA was tested for its quality using 1% agarose gel electrophoresis and the DNA samples were stored at –20°C until use.
Primer design and polymerase chain reaction
The coding region sequences of the PRL gene were retrieved from Ensembl Genome Browser (http://www.ensembl.org/Homo_sapiens) and the oligonucleotide primer sets flanking exons 1–5 were designed using Primer3plus (https://primer3plus.com/) software. Validation of primer sets was done using Primerstat(https://www.bioinformatics.org/sms2/pcr_primer_stats.html) and Blat (https://genome.ucsc.edu/cgi-bin/hgBlat) webservers. The best primer sets were later synthesized at a commercial facility (Table I). Each PCR reaction was carried out in a 25 µl reaction mixture using 1 µl of genomic DNA, 10 pmol of forward and reverse primers, 9.5 µl of distilled water and 12.5 µl of GoTaq Green Master Mix (Promega USA, Cat. #M7122) containing 1X PCR buffer, 1.5 mM magnesium chloride, 0.2 mM dNTPs, and 2 units of Taq DNA polymerase. PCR amplification included 3 steps, an initial denaturation at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 57.6°C for 30 s and primer extension at 72°C and a final extension step of 72°C for 5 min. After PCR reaction, the amplicons were electrophoresed on 2% agarose gel with size markers, stained with SYBRVR Safe Nucleic Acid Gel stain, (Invitrogen Inc.) and the bands were analyzed using UVitec Gel Documentation system-232 (UVitec, Cambridge, UK).
Table I
List of prolactin gene primer sets
Sanger sequencing, sequence alignment, and mutation identification
Initially, PCR products of the amplified exons (1–5) were purified using QIA quick PCR Purification Kit following the manufacturer’s instructions (Qiagen, Alameda, CA, USA). The purified PCR product was used as a DNA template for cycle sequencing reaction using the ABI 3500 Genetic Analyzer (Life Technologies, USA). Cycle sequencing PCR was carried out using 1 µl of big dye, 2 µl of 5X big dye buffer, 1 µl of either forward or reverse primer, and 1 µl of purified PCR product from 100–300 bp and 5 µl nuclease-free water. Cycle sequencing program conditions were as follows: denaturation at 96°C for 30 s, followed by 25 cycles of denaturation 95°C for 15 s, annealing at 57.6°C for 30 s and extension at 72°C for 2 min, and a final extension step of 72°C for 5 min. All the PCR products were analyzed using the ABI 3500 Genetic Analyzer.
BioEdit v6 software (www.mbio.ncsu.edu/BioEdit/bioedit.html) was used for aligning wild-type and sample sequences in order to identify the variants/mutations. Somatic mutations were sequence changes observed only in leiomyomas and not in their corresponding normal myometrium tissues, whereas germline variants were sequence changes seen in both fibroid and myometrium tissues.
Serum prolactin measurements
The prolactin in vitro Enzyme-Linked Immunosorbent Assay (ELISA) kit (Cat No. 4226-19, Cortez Diagnostics, California) was used for the quantitative measurement of prolactin levels of serum samples. Initially, to all the samples, standards and anti- prolactin-horseradish peroxidase enzyme (HRP) were added to a 96-well plate (precoated with anti-prolactin IgG antibodies) where the prolactin and anti-prolactin-HRP bound to the antibody to form a complex. The mixture was incubated at room temperature and then washed to remove any unbound material. Then, a tetramethylbenzidine (TMB) reagent was added, resulting in a blue color as it was catalyzed by HRP. Finally, a stop solution was added to terminate the reaction, causing the color to change from blue to yellow. The intensity of the signal is directly proportional to the amount of prolactin in the sample and the intensity was measured at 450 nm.
In-silico prediction
All the genetic variants were uploaded to the Variant Effect Predictor (VEP) tool, in Human Genome Variation Society (HGVS) format. The output data provided information about the gene, transcripts, variant location, consequence, minor allele frequency (MAF) in population groups and pathogenicity prediction scores generated by different computational webservers [25]. From the output data we selected the prediction results of Scale-Invariant Feature Transform (SIFT) [26], Polymorphism Phenotyping-2 (PolyPhen-2) [27], Combined Annotation Dependent Depletion (CADD) [28], and Functional Analysis through Hidden Markov Models (FATHMM) [29, 30] to define whether the observed variants are damaging, possibly damaging or benign to the function of the prolactin gene.
Statistical analysis
Analysis of serum prolactin levels between patient and control groups was performed using the GraphPad quick calc, v. 6.0 online statistics software (GraphPad Software Inc, USA). The data were expressed as mean ± standard deviation. The prolactin serum level was compared in patients and controls by the two-tailed t test. A statistically significant difference in serum levels was determined with a 95% confidence interval (95% CI) for odd’s ratio to a cut-off p-value of < 0.05.
Linkage disequilibrium analysis
The HaploReg webserver explores annotations of the noncoding genome at variants on haplotype blocks [31]. It includes linkage disequilibrium (LD) information from the 1000 Genomes Project, annotations from the Roadmap Epigenomics and the Encyclopedia of DNA Elements (ENCODE) projects, sequence conservation across mammals, effect of SNPs on regulatory motifs and on gene expression from eQTL studies. We used HaploReg (v. 4.1) to identify the tightly linked variants with r2 ≥ 0.8 and to examine their effect on gene function.
Results
Clinical analysis
Participants’ characteristics
Most of the patients were found to be of menopausal age (54%), whereas control participants were mostly of premenopausal age (59%). Regarding the BMI, 46% of patients were obese (Table II), while more than half (52%) of the control participants were overweight. Patients in this study presented at least one or two symptoms associated with UL; of these the most common were irregular uterine bleeding (46%) and anemia (24%) (Table III).
Table II
Age, menopausal state and anthropometric characteristics of UL patients
Table III
Clinical characteristics of UL patients
Leiomyoma characteristics
Histological classification showed that most tumors were the common leiomyoma type (82%) as opposed to cellular leiomyoma (6%) or mitotically active leiomyomas (3%). When investigating the location of leiomyomas, most were found to be intramural (IM) (52%), followed by subserosal (SS) (8%) and submucosal leiomyomas (SM) (4%). The remainder, however, were found to be in a heterogenous form (Table III). Moreover, analysis of the number of occurrence of leiomyomas revealed that multiple leiomyomas (≥ 2 nodes) were greater in number (40/50; 80%) in comparison to single (10/50; 20%) leiomyomas. The average size of leiomyomas was 5.3 ±4.5 cm, with a range of 0.43–13.3 cm.
Prolactin measurement
Out of 50 UL patients, 44 (88%) were reported to have high prolactin levels, with a mean value of 172.1 ±139.2 (Pmol/l). The prolactin levels of 50 control participants showed a mean value of 15.18 ±7.52 (Pmol/l). Statistical analysis showed a highly significant difference (p < 0.0001; OR [95% CI] = 156.9200 [117.7971–196.0429]) between groups, indicating a significant elevation of PRL serum level in UL patients.
PRL mutation analysis
The results of direct nucleotide sequencing of PRL gene exons revealed no evidence of somatic mutations in the fibroid tissues, ruling out their role in fibroid development. However, 4 variants – rs6239 (c.570G>A), rs16885980 (c.312+177T>C), rs1205955 (c.205-102T>A) and rs151282935 (c.269C>T) – were found in both fibroid and myometrium tissues (Figure 1). Variant c.570G>A was found in a heterozygous state (G/A) in 5 myometrium and 5 fibroid tissues and homozygous (A/A) in 2 myometrium and 2 fibroid tissues. Similarly, variant c.205-102T>A was found in the heterozygous state (T/A) in 20 myometrium tissues as well as 20 fibroid tissues and, homozygous (A/A) in 7 myometrium tissues and 7 fibroid tissues. Two variants (c.312+177T>C and c.269C>T) though, were only found in the heterozygous state. Variant c.312+177T>C was found in heterozygous state (T/C) in 5 myometrium tissues and 5 fibroid tissues and variant c.269C>T, was also found in heterozygous state (C/T) in 2 myometrium and 2 fibroid tissues respectively (Table IV). Among these 4 variants, 2 (c.269C>T) and c.570G>A were located in the coding region (exon 3 and exon 5) and 2 (c.205-102T>A and c.312+177T>C) in the non-coding region (intron 2 and intron 3). The MAF range for these variants is 0.01 to 0.26 in gnomAD and/or SHGP. Since these variants were found in both the leiomyoma and myometrium tissues, we conclude that they are germline polymorphisms.
Figure 1
ABI sequenced chromatograms showing the detected PRL variants. A – c.570 G>A, B – c.205-102 T>A, C – c.312+177 T>C, D – c.269 C>T
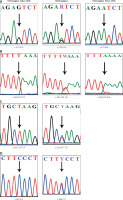
Table IV
PRL genetic variants identified in UL cases
Minor allele frequency analysis of PRL variants
The minor allele frequencies (MAF) of the identified variants were retrieved from different databases including the 1000 Genomes Project (1KGP), Saudi Human Genome Project (SHGP) and the Great Middle Eastern (GME) Variome Project. Variant. rs6239 was reported across different populations, whereas variant rs151282935 was seen only in the African population as well as being reported in the SHGP and GME (Table IV). Variant rs1205955 was reported in different populations but not the GME. Lastly, variant rs16885980 was seen in different populations and was not reported in the SHGP or GME (Table IV).
In-silico analysis of PRL variants
Table V shows the pathogenicity prediction scores for the four identified PRL variants. The c.269C>T variant was predicted as mostly damaging and possibly damaging by all 4 tools, SIFT, POLYPHEN2, CADD and FATHMM. For the remaining 3 variants (c.570G>A, c.205-102T>A and c.312+177T>C), only the CADD prediction tool predicted them as neutral or tolerated.
Table V
Pathogenicity prediction of variants
Linkage disequilibrium analysis of PRL variants
Linkage disequilibrium analysis of the 4 identified variants revealed that they are in total compliance with Hardy-Weinberg equilibrium. Variant rs6239 (c.570G>A) and rs16885980 (c.312+177T>C) are in strong linkage with each other and with third variant, rs75493272. Variant rs6239 alters the motifs of the CCNT2 gene, whereas variant rs16885980 alters the motifs of different transcription factors (TF) in the PRL gene. The rs1205955 variant is in strong linkage with rs1205960 (r2 ≥ 0.8) and similarly alters the motifs of different TFs. The last variant, rs151282935, potentially alters the TF motif of the STAT gene.
Discussion
Uterine leiomyoma is among the most frequent entities encountered in the practice of gynecology by the age of 50 [14]. There are three types of leiomyomas: intramural (most common), subserosal and submucosal leiomyomas [32, 33]. In this study, intramural tumors were the most common type observed, followed by subserosal tumors. Previous studies have reported that approximately 59.8% of patients with intramural fibroids suffer abnormal uterine bleeding [34]. Most of our patients reported to have abnormal uterine bleeding (46%) followed by anemia (24%). Additionally, another common symptom is pelvic pain, reported in more than 30% of UL patients [35]. In the present study however, only 9% of patients experienced pelvic pain. A possible explanation for this may be lack of patient information. Age is a risk factor contributing to the development of UL [36, 37]. Several studies suggest that the prevalence of fibroids increases with age: 60% by the age of 35 and 80% by the age of 50 [34]. In this study, most patients were found to be between 41 and 50 years old (54%), followed by > 51 years (28%) and only 18% of them were less than 40 years old. Similarly, obesity is another risk factor contributing to UL [36]. Previous study reported that most patients were obese (47.1%), followed by patients with normal weight (34.4%) and overweight (18.5%) [38]. Our data are similar in that most patients (46%) were obese; however, overweight (38%) participants outnumbered those of normal weight (16%). Moreover, our control group BMI showed that mostly participants were overweight (52%), followed by obese (26%), and the remaining were normal weight (22%). A possible explanation behind our participants being mostly obese and overweight is that obesity is seen to be increasing among the Saudi population in recent decades [39, 40].
In normal conditions, prolactin is involved in reproduction and homeostasis. In terms of reproduction, prolactin exerts multiple effects such as the development of mammary glands and the synthesis and maintenance of milk secretion. Prolactin ensures homeostasis by maintaining a constant internal environment through regulation of the immune system, osmatic balance, and angiogenesis [41]. Prolactin is produced both by the pituitary gland and uterine tissues, including the endometrium and myometrium. Previous studies have shown that PRL may act as a growth factor in human tumorigenesis in breast, prostate, and endometrium [42]. PRL is one of the most highly expressed genes among patients with leiomyomas, especially with HMGA2 subtype [3]. The present study has supported the previous study by reporting a statistically significantly higher level of prolactin in serum among UL patients compared to the control group. A study suggested the use of serum prolactin level as a marker to confirm the diagnosis of UL [43]. Leiomyomas have the ability to synthesize prolactin [44]. Additionally, prolactin levels in patients with UL before myomectomy or hysterectomy were significantly elevated when compared to controls [44]. The present study confirms that the serum prolactin level positively correlates with the number and size of leiomyoma.
No somatic mutations were detected in the PRL gene, thus ruling out its direct involvement in uterine tumorigenesis. Four genetic variants, i.e. rs6239, rs16885980, rs1205955 and rs151282935, detected in the PRL gene are in total compliance with Hardy-Weinberg equilibrium. Although a specific association of these four polymorphisms with UL is not yet reported, it is likely that these variants contribute to the fibroid development through the genes whose expression they control. For example, rs6239 and rs16885980 variants are in strong linkage with each other and with rs75493272 (r2 ≥ 0.8). The first variant (rs6239) was found to alter the motifs of the CCNT2 gene, located on chromosome 2, which plays a role in mitosis [26] and in regulating p53 activity, which is critically impaired in different cancers [29]. The second variant, rs16885980, has been found to disrupt the activity of different transcription factors (TF) in the PRL gene – Foxc1_2, Foxd1_, Maf_disc2, YY1_disc1, and YY1_known5 – which further regulates the expression many driver genes of cancers [45–47]. The third variant, rs1205955, is in strong linkage with variant rs1205960 (r2 ≥ 0.8) and has the ability to potentially alter the PRL gene transcription motifs such as AFP1, Mef2_known1, Mef2_known6, Ncx_2, Pax-4_2, Pou2f2_known8 and TATA_known. The fourth variant, rs151282935 (c.269C>T), has however been found to affect the TF motif called STAT, which is often overexpressed in various malignancies [48].
Based on the abovementioned indications, it is assumed that these and other germline polymorphisms could potentially influence the PRL gene activity through TF motifs, which further regulates different interacting genes. The altered PRL gene transcription could be the underlying cause of elevated levels of serum prolactin hormone in UL patients.
A possible explanation for the absence of damaging mutations in the PRL gene is that it is a highly conserved gene with a very crucial role in the survival of human beings in the very first years of infancy. Mutations in this gene would be threatening to survival. Hence, a possible hypothesis would be that due to natural selection and survival mechanisms damaging mutations have been eliminated over time.
Moreover, MAF scores of the identified variants further show the rarity of common variants in the PRL gene as well. All the variants, excluding variant rs1205955, were found to be either rare or not reported across different populations including the Saudi population. Variant rs1205955, however, was only rare in the Saudi population and not reported in the Middle Eastern database as opposed to other populations. This could be attributed to the high consanguinity rates in the Saudi and Arab populations.
This study sincerely, admits few limitations. This study only generated data from the coding regions of the PRL gene and did not cover the non-coding regions such as introns, 3ʹ and 5ʹ regions, which usually affect the transcript expression. Due to inevitable delays in obtaining fresh small tissue biopsies of tumor samples from the histopathology department, we could not measure the prolactin hormone in tissues. Our small sample size may not allow us to generalize our findings to UL. However, our study, being the first one reporting the lack of PRL gene somatic mutations, could act as a reference for future investigations.
In conclusion, this is the first study from Saudi Arabia to ascertain the elevated PRL serum levels in UL patients. This study has also confirmed the lack of evidence for the role of somatic mutations in the PRL gene in fibroids. This study detected 4 germline variants in the PRL gene whose exact association with fibroid development remains unclear. Hence, we recommend a larger case-control study to verify whether PRL gene germline polymorphisms could act as potential genetic markers for the disease diagnosis, prognosis and/or its treatment.


