Introduction
One of the leading causes of cancer-related death is lung cancer, in which non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung-cancer-caused mortality [1, 2]. The NSCLC patients can obtain remission with modern therapies, such as radio-chemotherapy, immunotherapy, epidermal growth factor receptor tyrosine kinase inhibitors (EGFR TKIs) therapy and ALK rearrangements [2–5]. But the 5-year survival rate for patients with metastatic NSCLC remains extremely poor [6]. Some studies have indicated that non-coding RNAs (ncRNAs) could act as regulators in the development and as biomarkers in the treatment of NSCLC [7, 8]. Hence, investigation of the mechanism of more novel ncRNAs in NSCLC is urgently needed.
Circular RNAs (circRNAs), a class of highly conserved ncRNAs, lack 5’ and 3’ polarity. The covalently closed-loop structures endow circRNAs with low degradation and high stability [9, 10]. The involvement of circRNAs in human cancers has been gradually recognized. Guan et al. asserted that circPUM1 was conductive to the tumorigenesis of ovarian cancer acting as sponges of miR-615-5p and miR-6753-5p [11]. Wang et al. discovered that the up-regulation of hsa_circ_0000517 could serve as a prognostic marker in hepatocellular carcinoma [12]. Liu et al. reported that hsa_circ_103809 expedited the progression of lung cancer via the miR-4302/ZNF121/MYC axis [13]. A recent report showed abnormally high expression of circ_0001421 in NSCLC tissues [14], while the role of circ_0001421 in NSCLC is unknown.
As other frequent ncRNAs, miRNAs usually participate in varied biological behaviors and the development of tumors as pivotal regulators [15, 16]. MiR-221 was shown to function as a tumor promoter in NSCLC by targeting TIMP2 [17]. Ma et al. proved the antitumor role of miR-223 in NSCLC via the EGFR-mediated PI3K/AKT signal pathway [18]. Song et al. reported that miR-409 restricted NSCLC progression by regulating SPIN1 [19]. Also miR-409-3p was found to be involved in the NSCLC progression regulated by long ncRNA (lncRNA) ZEB1-AS1 [20]. Herein, we intended to explore the influence of miR-409-3p on NSCLC and the relation with circ_0001421.
Transmembrane protein 14A (TMEM14A) belongs to the TMEM family, and plays an essential role in several cancers. For example, the knockdown of TMEM14A was manifested to decrease cell proliferation and invasion in ovarian cancer [21]. An et al. reported that TMEM14A was overexpressed and regulated by the circ_0003645/miR-1179 axis in NSCLC [22]. Our study focused on the connection among circ_0001421, miR-409-3p and TMEM14A in the regulation of cell colony formation, migration, invasion and glycolysis of NSCLC.
Material and methods
Tissues source and cell culture
After obtaining the informed consent from NSCLC patients, a total of 38 pairs of NSCLC tissues and normal peritumor tissues were acquired at Xiantao First People’s Hospital Affiliated to Changjiang University, followed by the preservation of samples in liquid nitrogen. These patients did not receive any other therapy prior to pneumonectomy. Our research was authorized by the Institute Review Ethics Committee of Xiantao First People’s Hospital Affiliated to Changjiang University.
Human NSCLC cell lines (H1975 and A549) and the normal lung epithelial BEAS-2B cell line from American Type Culture Collection (ATCC, Manassas, VA, USA) were cultivated in the environment of 37°C, 5% CO2 and 95% air. The culture medium consisted of the basic medium Roswell Park Memorial Institute-1640 (RPMI-1640; Life Technologies, Carlsbad, CA, USA), 10% fetal bovine serum (FBS; Serapro, Naila, Germany) and 1% penicillin-streptomycin (Gibco, Carlsbad, CA, USA).
Transient transfection
Small interfering RNA (siRNA) against circ_0001421 (si-circ_0001421), miR-409-3p mimic and inhibitor (miR-409-3p and anti-miR-409-3p) were synthesized by GENEWIZ (Suzhou, China). Vector construction was implemented by cloning the open reading frame (ORF) of TMEM14A into the pcDNA3.0 basic vector (Invitrogen, Carlsbad, CA, USA) to overexpress TMEM14A (TMEM14A). Transiently, these oligonucleotides or vector were transfected into NSCLC cells following the Lipofectamine 3000 (Invitrogen) protocol. Si-NC, miR-NC, anti-miR-NC and vector were used as the negative controls for the above transfection.
Quantitative real-time polymerase chain reaction
The RNA extraction was executed using Trizol (Invitrogen), and 2 µg RNA was used to synthesize complementary DNA (cDNA) with the PrimeScript RT Master Mix (Takara, Dalian, China), then quantitative real-time polymerase chain reaction (qRT-PCR) reaction was conducted using a Fast SYBR Green PCR kit (Applied Biosystems, Foster City, CA, USA). We used glyceraldehyde-3-phosphate dehydrogenase (GAPDH, for circ_0001421 and TMEM14A) and U6 (for miR-409-3p) as internal controls to normalize the expression levels. Primer sequences were as follows: circ_0001421 (forward: 5’-TGCCCAGAATGACAAGCATA-3’ and reverse: 5’-TGAGCAGATGTTCCTGTTGC-3’); miR-409-3p (forward: 5’-GGGGAATGTTGCTCGGTGA-3’ and reverse: 5’-CAGTGCGTGTCGTGGAGT-3’); TMEM14A (forward: 5’-GTTTGGTTGCAGGTTTAAG-3’ and reverse: 5’-AATATGCTCTGCCCATTAC-3’); GAPDH (for-ward: 5’-GTCTCCTCTGACTTCAACAGCG-3’ and reverse: 5’-ACCACCCTGTTGCTGTAGCCAA-3’); U6 (for-ward: 5’-CTCGCTTCGGCAGCACA-3’ and reverse: 5’-AACGCTTCACGAATTTGCGT-3’). Eventually, the data were exhibited as relative expression levels by the 2-∆∆Ct method [23].
Localization analysis
The supernatant from centrifugal cell lysate was considered as the cytoplasmic fraction and the pellet was the nucleus fraction. The PARIS Kit (Invitrogen) was applied for isolating RNA from the nuclear and cytoplasmic fractions, referring to the user’s guideline. The expression levels of circ_0001421, GAPDH and U6 were detected by qRT-PCR using two RNA as samples. GAPDH and U6 were separately used as the positive controls of cytoplasmic and nuclear fractions.
Colony formation assay
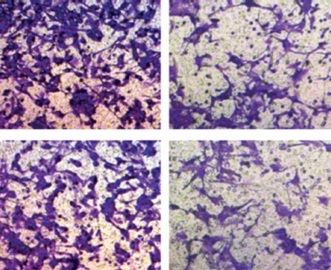
Transfected NSCLC cells were plated into 6-well plates with 1000 cells per well. Following inoculation about 2 weeks, macroscopic colonies were fixed using 4% paraformaldehyde and dyed with crystal violet (Sangon, Shanghai, China). After washing with phosphate buffer solution (PBS; Corning, New York, NY, USA), the counting of colonies was performed with a microscope.
Detection of migration and invasion
Transwell assay was administered to assess cell migration and invasion abilities. Concisely, a 100 µl cell suspension was pipetted into the upper chamber of a transwell chamber (Corning) and 600 µl of culture medium was added to the lower chamber, incubating for 24 h. After the respective fixing and staining with 4% paraformaldehyde and crystal violet (Sangon), the migrated cells were counted under a microscope. But in the detection of invasion, the upper chamber must be coated with Matrigel (Corning) before cell seeding, and invaded cells were calculated.
Measurement of the extracellular acidification rate
The Seahorse Extracellular Flux Analyzer XF96 (Seahorse Bioscience, Billerica, MA, USA) was used for measuring the extracellular acidification rate (ECAR) in vitro, complying with the manufacturer’s protocol. Briefly, transfected NSCLC cells (2 × 104 cells/well) were inoculated into an XF96-well plate to attach for 16–20 h under the culture conditions. After that, cell medium was replaced with XF assay unbuffered medium complemented with 2 mM glutamine, followed by the injection of 10 mM glucose, 1 µM oligomycin, and 80 mM 2-deoxyglucose. The measured ECAR was recorded as mpH/min.
Assessment of glucose consumption and lactate production
Non-small cell lung cancer cells were seeded into 24-well plates overnight. After cell transfection, the glucose consumption and lactate production levels were assayed from cell lysates via the glucose uptake colorimetric assay kit and lactate colorimetric assay kit (BioVision, Milpitas, CA, USA), respectively, according to the operating procedures [24].
Western blot
The protein was extracted using radio immunoprecipitation assay (RIPA) lysis buffer (Sangon), and the separation of 40 µg proteins was performed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by transfer onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA) and blockage by 5% non-fat milk (Sigma, St. Louis, MO, USA). The membranes were incubated with primary antibodies: anti-glucose transporter 1 (anti-GLUT1; Abcam, Cambridge, UK, ab115730, 1 : 1000), anti-lactic dehydrogenase (anti-LDHA; Abcam, ab101562, 1 : 1000), anti-TMEM14A (Sigma, HPA072536, 1 : 1000), internal control anti-GAPDH (Abcam, ab181602, 1 : 3000) for 4 h at indoor temperature. Following the incubation of secondary antibody (Abcam, ab205718, 1 : 5000) for 1 h, the presence of immunoconjugated signals was conducted by the enhanced chemiluminescence reagent (Millipore), then Image Lab software version 4.1 (Bio-Rad Laboratories, Hercules, CA, USA) was used for collecting images and analyzing signal density [25].
Dual-luciferase reporter assay
The amplified circ_0001421 sequences, including wild-type (WT, containing the putative miR-409-3p sites) and mutant-type (MUT, containing the mutant miR-409-3p sites), were constructed into psiCHECK-2 luciferase basic vector (Promega, Madison, WI, USA) to obtain the recombinant luciferase reporters circ_0001421 WT and circ_0001421 MUT. The 3’-UTRs of TMEM14A WT and MUT were also fused to acquire TMEM14A 3’-UTR WT and TMEM14A 3’-UTR MUT. NSCLC cells were transfected with the constructs and miR-409-3p or miR-NC for 48 h, and cell lysate by the passive buffer (Promega) was used for examination of the luciferase intensity by the dual-luciferase reporter system (Promega), abiding by the producer’s manual. The relative luciferase activity was expressed as the ratio of firefly/Renilla luciferase intensity.
Xenotransplantation assay
The A549 cells stably expressing sh-circ_0001421 were constructed through the transfection of lentivirus vector containing short hairpin RNA (shRNA) targeting circ_0001421 (sh-circ_0001421; GenePharma, Shanghai, China), using sh-NC as the negative control. Vital River Laboratory Animal Technology (Beijing, China) provided the purchase service of male BALB/c nude mice (n = 10, 6-week-old) for us. Then mice underwent subcutaneous injection with transfected A549 cells (1 × 106/50 µl PBS), and were divided into sh-circ_0001421 and sh-NC groups (5 mice per group). Tumor burden was monitored through digital calipers weekly, and the acknowledged formula (length × width2 × 0.5) was applied for estimating tumor volume. All mice were subjected to euthanasia 5 weeks after injection, then tumors were collected and the weight was measured using an electronic scale. Ultimately, the levels of circ_0001421, miR-409-3p and TMEM14A in the tumors were detected. The animal assay received permission from the Animal Ethics Committee of Xiantao First People’s Hospital Affiliated to Changjiang University.
Statistical analysis
Independently, the experiments in our study were repeated three times. Data were expressed as the mean ± standard deviation (SD). Statistical analyses were performed using SPSS 20.0 and graphic plotting was conducted through GraphPad Prism 7. Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were implemented for the difference analysis. A value of p < 0.05 was considered statistically significant.
Results
Circ_0001421 was aberrantly overexpressed in NSCLC tissues and cells
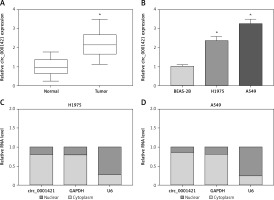
qRT-PCR analysis was used to affirm whether circ_0001421 was dysregulated in NSCLC. As Figure 1 A illustrates, the circ_0001421 level was much higher in NSCLC tissues than that in normal tissues. Also, the overexpression of circ_0001421 was validated in H1975 and A549 cells by contrast to normal BEAS-2B cells (Figure 1 B). And interestingly, both circ_0001421 and GAPDH were abundant in cytoplasm but small nuclear RNA U6 existed in the nucleus principally, suggesting the location of circ_0001421 in cytoplasm (Figures 1 C, D). Mainly, we verified that circ_0001421 was overexpressed in NSCLC.
Knockdown of circ_0001421 restrained colony formation, migration, invasion and glycolysis of NSCLC cells
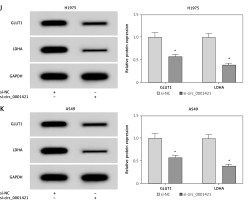
si-circ_0001421 transfection was applied to knock down circ_0001421 for researching the role of circ_0001421 in NSCLC cells. QRT-PCR revealed the successful interference of si-circ_0001421 with circ_0001421 expression compared to the si-NC group (Figures 2 A, B). Subsequently, the further function exploration was implemented. In colony formation assay, fewer colonies were observed in the si-circ_0001421 group than the si-NC group (Figure 2 C). Transwell assay showed that the number of migrated (Figure 2 D) and invaded (Figure 2 E) cells remarkably declined following the down-regulation of circ_0001421. The indicators of glycolysis are numerous, such as ECAR, glucose consumption, lactate production and the glycolysis-related proteins including glucose transporter-1 (GLUT1) and lactate dehydrogenase A (LDHA) [26, 27]. The experimental results indicated that circ_0001421 knockdown decreased the ECAR in both H1975 (Figure 2 F) and A549 (Figure 2 G) cells, as well as the glucose consumption (Figure 2 H) and lactate production (Figure 2 I). Western blot revealed that the protein expression levels of GLUT1 and LDHA were repressed in H1975 (Figure 2 J) and A549 (Figure 2 K) cells transfected with si-circ_0001421. Altogether, circ_0001421 down-regulation exerted inhibitory effects on colony formation, migration, invasion and glycolysis in NSCLC cells.
Figure 2
Knockdown of circ_0001421 restrained colony formation, migration, invasion and glycolysis of NSCLC cells. H1975 and A549 cells were transfected with si-circ_0001421 or si-NC. A, B – The knockdown efficiency of si-circ_0001421 in H1975 and A549 cells was analyzed by qRT-PCR. C – The colony formation ability was detected using colony formation assay. D, E – Transwell assay was used for assessing cell migration and invasion D, E – Transwell assay was used for assessing cell migration and invasion. F–K – The glycolysis was estimated through ECAR measurement (F, G), glucose consumption (H), lactate production (I) The protein levels of GLUT1 and LDHA by Western blot (J, K). *P < 0.05
Circ_0001421 served as a sponge of miR-409-3p
Increasing studies have testified that circRNAs could act as miRNA “sponges” in various human cancers [28–30]. After the prediction of StarBase v2.0 software, we noted that circ_0001421 contained the binding sites of miR-409-3p (Figure 3 A). The interaction between circ_0001421 and miR-409-3p was certified by dual-luciferase reporter assay. As shown in Figures 3 B, C, miR-409-3p co-transfection with circ_0001421 WT strikingly reduced the relative luciferase activity of H1975 and A549 cells, but there was no conspicuous difference in luciferase activity after miR-409-3p co-transfection with circ_0001421 MUT. In comparison to normal tissues and BEAS-2B cells, miR-409-3p expression was significantly reduced in both NSCLC tissues (Figure 3 D) and cells (Figure 3 E). And notably, miR-409-3p level was up-regulated in H1975 (Figure 3 F) and A549 (Figure 3 G) cells transfected with si-circ_0001421, which implied that circ_0001421 acted as a miR-409-3p sponge.
Figure 3
Circ_0001421 served as a sponge of miR-409-3p. A – The bioinformatic analysis between circ_0001421 and miR-409-3p was performed by StarBase v2.0. B, C – The correlation between circ_0001421 and miR-409-3p was analyzed using the dual-luciferase reporter assay. D, E – The level of miR-409-3p in NSCLC tissues and cells was detected through qRT-PCR. F, G – The qRT-PCR was applied for assessing the influence of circ_0001421 inhibition on the expression of miR-409-3p in NSCLC cells. *P < 0.05
Inhibition of miR-409-3p reverted the impacts of circ_0001421 knockdown on NSCLC cells
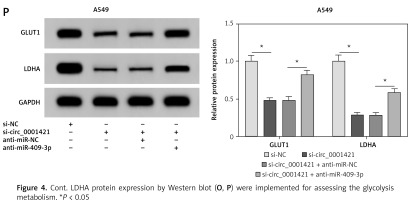
Next, we used rescue experiments to study the regulatory relation between circ-0001421 and miR-409-3p. qRT-PCR demonstrated that anti-miR-409-3p transfection significantly recovered the si-circ_0001421-induced increase of miR-409-3p expression, suggesting that the inhibitory effect of anti-miR-409-3p on miR-409-3p level was excellent (Figures 4 A, B). In addition, the repression of colony formation (Figures 4 C, D), migration (Figures 4 E, F) and invasion (Figures 4 G, H) caused by circ_0001421 depression in H1975 and A549 cells were all prominently reversed following the inhibition of miR-409-3p level. Seahorse analysis showed that the si-circ_0001421-motivated suppressive effect on ECAR was reversed by miR-409-3p inhibitor (Figures 4 I, J). Also, si-circ_0001421 and anti-miR-409-3p co-transfection enhanced the glucose consumption (Figures 4 K, L) and lactate production (Figures 4 M, N) contrasted to the si-circ_0001421 + anti-miR-NC group. And the introduction of anti-miR-409-3p offset the down-regulation of GLUT1 and LDHA protein expression evoked by the low expression of circ_0001421 in H1975 (Figure 4 O) and A549 (Figure 4 P) cells. All the data proved that the impacts of circ_0001421 knockdown on NSCLC cells were relieved through inhibiting miR-409-3p.
Figure 4
Inhibition of miR-409-3p reversed the impacts of circ_0001421 knockdown on NSCLC cells. H1975 and A549 cells were transfected with si-NC, si-circ_0001421, si-circ_0001421 + anti-miR-NC or si-circ_0001421 + antimiR- 409-3p. A, B – miR-409-3p expression was determined using qRT-PCR. C, D – Colony formation assay was performed to evaluate the colony ability of cells. E–H – Cell migration (E, F). Invasion (G, H) were analyzed by transwell assay. I–P – The ECAR detection (I, J), glucose consumption (K, L). Lactate production (M, N) and the GLUT1 and LDHA protein expression by Western blot (O, P) were implemented for assessing the glycolysis metabolism. LDHA protein expression by Western blot (O, P) were implemented for assessing the glycolysis metabolism. *P < 0.05.
TMEM14A was a target of miR-409-3p and circ_0001421 regulated TMEM14A expression by sponging miR-409-3p
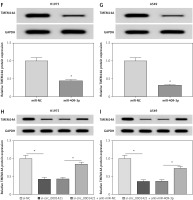
MiRNAs can usually interact with the 3’UTR of downstream gene [31, 32]. StarBase v2.0 displayed the binding sites between TMEM14A 3’UTR and miR-409-3p (Figure 5 A). Furthermore, the luciferase activity of the TMEM14A 3’UTR WT group was refrained by miR-409-3p overexpression, while it remained unchanged in the TMEM14A 3’UTR MUT group (Figures 5 B, C). After verifying the combination of miR-409-3p and TMEM14A, we measured the expression of TMEM14A in NSCLC. Distinctly, NSCLC tissues (Figure 5 D) and cells (Figure 5 E) expressed a rising tendency of TMEM14A protein level, compared to normal tissues and cells. After miR-409-3p was transfected into H1975 (Figure 5 F) and A549 (Figure 5 G) cells, the decrease of TMEM14A protein expression was presented by Western blot. Moreover, transfection of si-circ_0001421 triggered the down-regulation of TMEM14A, which was partly rescued through miR-409-3p depletion (Figures 5 H, I). These results indicated that miR-409-3p targeted TMEM14A and circ_0001421 modulated TMEM14A via sponging miR-409-3p.
Figure 5
TMEM14A was a target of miR-409-3p and circ_0001421 regulated TMEM14A expression by sponging miR-409-3p. A – The binding sites of miR-409-3p and TMEM14A were analyzed by StarBase v2.0. B, C – Dualluciferase reporter assay was used for confirming the combination between miR-409-3p and TMEM14A. D, E – TMEM14A protein expression in NSCLC tissues and cells was examined by Western blot. F, G – Western blot was applied to assess the effect of miR-409-3p on TMEM14A expression with miR-NC as the negative control. H, I – The protein level of TMEM14A was detected in H1975 and A549 cells transfected with si-circ_0001421, si-circ_0001421 + anti-miR-409-3p or matched controls. *P < 0.05
MiR-409-3p overexpression suppressed colony formation, migration, invasion and glycolysis via targeting TMEM14A in NSCLC cells
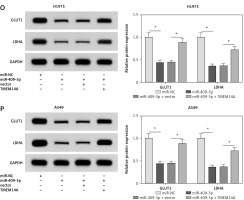
H1975 and A549 cells were transfected with miR-NC, miR-409-3p, miR-409-3p + vector or miR-409-3p + TMEM14A to research whether TMEM14A was responsible for the tumor inhibitor role of miR-409-3p in NSCLC. Firstly, TMEM14A was successfully overexpressed by TMEM14A transfection, which alleviated the miR-409-3p-induced restraint of TMEM14A protein expression (Figures 6 A, B). The overexpression of miR-409-3p resulted in the reduction of the number of cell colonies (Figures 6 C, D), migration (Figures 6 E, F) and invasion (Figures 6 G, H), whereas ectopic high expression of TMEM14A neutralized these effects. Additionally, the miR-409-3p-induced repressive influence on ECAR was abrogated following TMEM14A up-regulation in part (Figures 6 I, J). Likewise, miR-409-3p transfection evoked the inhibition of glucose consumption (Figures 6 K, L) and lactate production (Figures 6 M, N) as well as the protein levels of GLUT1 and LDHA (Figures 6 O, P), while this suppression was counteracted by the promotion of TMEM14A. We concluded that the tumor inhibitory function of miR-409-3p in NSCLC was achieved by targeting TMEM14A.
Figure 6
MiR-409-3p overexpression suppressed colony formation, migration, invasion and glycolysis via targeting TMEM14A in NSCLC cells. H1975 and A549 cells were transfected with miR-NC, miR-409-3p, miR-409-3p + vector or miR-409-3p + TMEM14A. A, B – Western blot was conducted for the determination of TMEM14A protein level. C, D – The detection of colony ability was performed using the colony formation assay. E–H – The evaluation of cell migration (E, F) and invasion (G, H) was administered through the transwell assay E–H – The evaluation of cell migration (E, F) and invasion (G, H) was administered through the transwell assay. I–P – The assessment of glycolysis was performed by ECAR measurement (I, J), glucose consumption (K, L), lactate production (M, N) and the associated protein (GLUT1 and LDHA) examination via Western blot (O, P). The associated protein (GLUT1 and LDHA) examination via Western blot (O, P). *P < 0.05
Depression of circ_0001421 reduced tumor growth of NSCLC via regulation of the miR-409-3p/TMEM14A axis in vivo
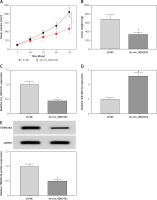
The effect of circ_0001421 on NSCLC in vivo was investigated by xenotransplantation assay. We observed the evident decrease of tumor volume (Figure 7 A) and weight (Figure 7 B) in the sh-circ_0001421 group, by comparison with the sh-NC group. Also, knockdown of circ_0001421 clearly reduced the expression of circ_0001421 (Figure 7 C) but enhanced the miR-409-3p level (Figure 7 D) in excised tumors. Western blot indicated that the protein expression of TMEM14A was repressed by inhibiting circ_0001421 in vivo (Figure 7 E). Taken together, circ_0001421 knockdown inhibited tumor growth of NSCLC by regulating the miR-409-3p/TMEM14A axis in vivo.
Figure 7
Depression of circ_0001421 reduced tumor growth of NSCLC via regulation of the miR- 409-3p/TMEM14A axis in vivo. A – Tumor volume was estimated weekly post-injection with A549 cells expressed sh-circ_0001421 or sh-NC. B – The weight of excised tumors was measured. C, D – The expression levels of circ_0001421 and miR-409- 3p were examined by qRT-PCR in tumors. E – The protein level of TMEM14A was determined using Western blot. *P < 0.05
Discussion
The molecular targeted therapy has been developed as a vital strategy to ameliorate the treatment and prognosis of NSCLC patients [33, 34]. In the present study, we found the overexpression of circ_0001421 and TMEM14A as well as the down-regulation of miR-409-3p in NSCLC. Circ_0001421 could promote NSCLC development by acting as a miR-409-3p sponge to promote expression of TMEM14A, suggesting that circ_0001421 has the potential to become a molecular target in NSCLC treatment.
Currently, the circRNAs have drawn the attention of many researchers in NSCLC. CircRNA SMARCA5 repressed the proliferation, migration, and invasion in NSCLC cells via regulation of the miR-19b-3p/HOXA9 axis [35]. Hsa_circ_0001946 had inhibitory effects on cell viability, migration and invasion of NSCLC through the nucleotide excision repair pathway [36]. Gao et al. attested that circ_0074027 elevated the proliferation and invasion abilities but reduced apoptosis by miR-185-3p/BRD4/MADD signals in NSCLC [37], and circRNA ZFR improved the NSCLC progression by the miR-101-3p/CUL4B axis [38]. These studies implied different roles of circRNAs in NSCLC. Here, circ_0001421 was verified to be up-regulated in NSCLC, and knockdown of circ_0001421 exhibited the suppressive impacts on cell colony formation, migration and invasion. Glycolysis, a biochemically oxygen-independent metabolic pathway, can act as a target in tumor therapy [39, 40]. Circ_0001421 down-regulation led to inhibition of glycolysis (ECAR, glucose consumption, lactate production and GLUT1 and LDHA protein levels). The oncogenic role of circ_0001421 was ascertained in NSCLC for the first time.
Regarding the functional mechanism behind circ_0001421 in NSCLC, we found the miRNA target of circ_0001421 and validated that circ_0001421 targeted miR-409-3p. According to the previous studies, miR-409-3p plays an anticancer role in the majority of cancers. For instance, miR-409-3p inhibited osteosarcoma cell proliferation and invasion via directly regulating ZEB1 [41]. Zhang et al. observed the down-regulation of miR-409-3p in breast cancer and miR-409-3p hindered cell growth and invasion via inhibiting Akt1 [42]. In line with this, rescue assays demonstrated that miR-409-3p up-regulation was responsible for the suppression of circ_0001421 knockdown in NSCLC progression, suggesting the suppressive role of miR-409-3p in NSCLC. Moreover, TMEM14A was a downstream target of miR-409-3p and miR-409-3p acted as the tumor inhibitor in NSCLC by targeting TMEM14A. Additionally, it is interesting that circ_0001421 positively modulated TMEM14A expression by functioning as a sponge of miR-409-3p. Judging from these results, we concluded that the effects of circ_0001421 on NSCLC cells were achieved by the miR-409-3p/TMEM14A axis. The circRNA-miRNA-mRNA regulatory network has been presented in multiple cancers [43–45]. Our experiments in vivo also suggested that circ_0001421 enhanced tumor growth through the miR-409-3p/TMEM14A axis.
Altogether, this study showed that circ_0001421 supported cell colony formation, migration, invasion and glycolysis in NSCLC via regulating the miR-409-3p/TMEM14A axis. The oncogenic role of circ_0001421 in NSCLC was revealed for the first time and the circ_0001421/miR-409-3p/TMEM14A axis afforded an unambiguous molecular pathomechanism for NSCLC, which might contribute to the understanding of the initiation and development of NSCLC in the circRNA field. Our findings indicated that circ_0001421 could improve the treatment of NSCLC as a novel molecular therapeutic target.