Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
DERMATOLOGY / CLINICAL RESEARCH
Identification of potential drug target genes for atopic dermatitis leveraging Mendelian randomization, single-cell transcriptomics, and spatial transcriptomics
1
Department of Dermatology, The Calmette Affiliated Hospital of Kunming Medical University, Kunming, China
2
Department of Dermatology, Kunming City Maternal and Child Health Hospital, Wuhua District, Kunming, China
3
Department of Dermatology, Guiyang Second People’s Hospital, Guanshanhu District, Guiyang, China
These authors had equal contribution to this work
Submission date: 2025-04-15
Final revision date: 2025-07-24
Acceptance date: 2025-07-31
Online publication date: 2025-10-05
Corresponding author
Qi Li
Department of Dermatology The Calmette Affiliated Hospital of Kunming Medical University Kunming, China Phone: +86-13698760791
Department of Dermatology The Calmette Affiliated Hospital of Kunming Medical University Kunming, China Phone: +86-13698760791
KEYWORDS
atopic dermatitisdrug targetMendelian randomizationsingle-cell RNA sequencingspatial transcriptomics
TOPICS
ABSTRACT
Introduction:
Atopic dermatitis (AD), the most common chronic inflammatory dermatosis, currently lacks definitive curative treatments. This study aimed to identify potential drug targets for AD through an integrative genomic approach.
Material and methods:
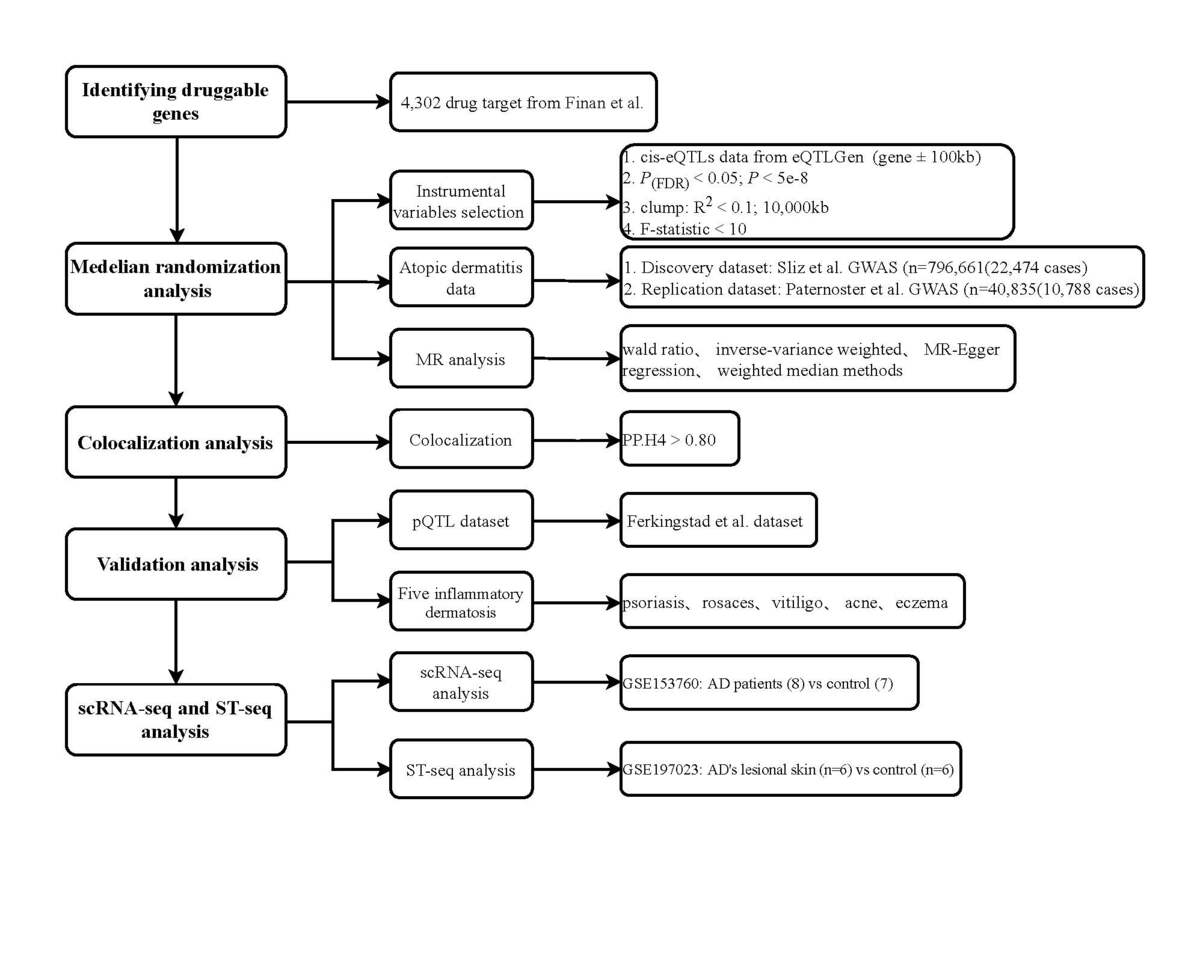
Cis-expression quantitative trait loci (cis-eQTL) from the eQTLGen consortium were used as genetic instruments for druggable genes. Summary-level AD statistics were obtained from the largest available GWAS dataset (cases = 22,474; controls = 774,187) with replication in an independent cohort (cases = 10,788; controls = 30,047). Mendelian randomization (MR) was employed to explore the causal relationship between druggable genes and AD risk, augmented by colocalization analysis to identify shared causal variants. A pQTL dataset was thereafter used for further validation. Furthermore, the potential association between the identified genes and five other inflammatory skin diseases was also assessed. Finally, we specifically investigated expression patterns of identified genes through analysis of single-cell RNA sequencing and spatial transcriptomics data from GEO datasets via Seurat.
Results:
Three druggable genes, HSP90AA1, IL2RA, and MANBA, were positively associated with an increased risk of AD. Colocalization analysis identified rs61839660 as a shared variant between IL2RA and AD, with pQTL data confirming IL2RA protein-level effects. Increased IL2RA gene expression was observed in natural killer cells within leukocyte infiltration regions. Moreover, MR analysis indicated that IL2RA gene expression also increases the risk of psoriasis and eczema, though without colocalization evidence.
Conclusions:
These findings suggest that IL2RA inhibitors could be promising therapeutic agents for the treatment of AD.
Atopic dermatitis (AD), the most common chronic inflammatory dermatosis, currently lacks definitive curative treatments. This study aimed to identify potential drug targets for AD through an integrative genomic approach.
Material and methods:
Cis-expression quantitative trait loci (cis-eQTL) from the eQTLGen consortium were used as genetic instruments for druggable genes. Summary-level AD statistics were obtained from the largest available GWAS dataset (cases = 22,474; controls = 774,187) with replication in an independent cohort (cases = 10,788; controls = 30,047). Mendelian randomization (MR) was employed to explore the causal relationship between druggable genes and AD risk, augmented by colocalization analysis to identify shared causal variants. A pQTL dataset was thereafter used for further validation. Furthermore, the potential association between the identified genes and five other inflammatory skin diseases was also assessed. Finally, we specifically investigated expression patterns of identified genes through analysis of single-cell RNA sequencing and spatial transcriptomics data from GEO datasets via Seurat.
Results:
Three druggable genes, HSP90AA1, IL2RA, and MANBA, were positively associated with an increased risk of AD. Colocalization analysis identified rs61839660 as a shared variant between IL2RA and AD, with pQTL data confirming IL2RA protein-level effects. Increased IL2RA gene expression was observed in natural killer cells within leukocyte infiltration regions. Moreover, MR analysis indicated that IL2RA gene expression also increases the risk of psoriasis and eczema, though without colocalization evidence.
Conclusions:
These findings suggest that IL2RA inhibitors could be promising therapeutic agents for the treatment of AD.
REFERENCES (37)
3.
Schuler CF, Billi Ac, Maverakis E, Tsoi LC, Gudjonsson JE. Novel insights into atopic dermatitis. J Allergy Clin Immunol 2023; 151: 1145-54.
4.
Hinkson IV, Madej B, Stahlberg EA. Accelerating therapeutics for opportunities in medicine: a paradigm shift in drug discovery. Front Pharmacol 2020; 11: 770.
5.
Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. Br J Pharmacol 2011; 162: 1239-49.
6.
Kiriiri GK, Njogu PM, Mwangi AN. Exploring different approaches to improve the success of drug discovery and development projects: a review. Future J Pharm Sci 2020; 6: 27.
7.
Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol 2014; 32: 40-51.
8.
Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet 2015; 47: 856-60.
9.
Smith GD, Ebrahim S, ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003; 32: 1-22.
10.
Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA 2017; 318: 1925-6.
11.
Chen Y, Xu X, Wang L, et al. Genetic insights into therapeutic targets for aortic aneurysms: a Mendelian randomization study. EBioMedicine 2022; 83: 104199.
12.
Lin J, Zhou J, Xu Y. Potential drug targets for multiple sclerosis identified through Mendelian randomization analysis. Brain 2023; 146: 3364-72.
13.
Chen J, Xu F, Ruan X, et al. Therapeutic targets for inflammatory bowel disease: proteome-wide Mendelian randomization and colocalization analyses. EBioMedicine 2023; 89: 104494.
14.
Chauquet S, Zhu Z, O’Donovan MC, et al. Association of antihypertensive drug target genes with psychiatric disorders: a Mendelian randomization study. JAMA Psychiatry 2021; 78: 623-31.
15.
Henry A, Gordillo-Maranon M, Finan C, et al. Therapeutic targets for heart failure identified using proteomics and mendelian randomization. Circulation 2022; 145: 1205-17.
16.
Yuan S, Wang L, Zhang H, et al. Mendelian randomization and clinical trial evidence supports TYK2 inhibition as a therapeutic target for autoimmune diseases. EBioMedicine 2023; 89: 104488.
17.
Chen L, Peters JE, Prins B, et al. Systematic Mendelian randomization using the human plasma proteome to discover potential therapeutic targets for stroke. Nat Commun 2022; 13: 6143.
18.
Finan C, Gaulton A, Kruger FA, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med 2017; 9: eaag1166.
19.
Vosa U, Claringbould A, Westra HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet 2021; 53: 1300-10.
20.
Sliz E, Huilaja L, Pasanen A, et al. Uniting biobank resources reveals novel genetic pathways modulating susceptibility for atopic dermatitis. J Allergy Clin Immunol 2022; 149: 1105-12 e9.
21.
Paternoster L, Standl M, Waage J, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet 2015; 47: 1449-56.
22.
Kurki MI, Karjalainen J, Palta P, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 2023; 613: 508-18.
23.
Jin Y, Andersen G, Yorgov D, et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat Genet 2016; 48: 1418-24.
24.
Palmer TM, Lawior DA, Sterne JAC, et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res 2012; 21: 223-42.
25.
Hartwig FP, Davies NM, Hemani G, Smith GD. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol 2016; 45: 1717-26.
26.
Teumer A. Common methods for performing mendelian randomization. Front Cardiovasc Med 2018; 5: 51.
27.
Burgess S, Scott RA, Timpson NJ, et al. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 2015; 30: 543-52.
28.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013; 37: 658-65.
29.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015; 44: 512-25.
30.
Bowden J, Smith GD, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016; 40: 304-14.
31.
Mitamura Y, Eiger M, Kim J, et al. Spatial transcriptomics combined with single-cell RNA-sequencing unravels the complex inflammatory cell network in atopic dermatitis. Allergy 2023; 78: 2215-31.
32.
Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 2013; 38: 13-25.
33.
Robert C, Kupper TS. Inflammatory skin diseases, T cells, and immune surveillance. N Engl J Med 1999; 341: 1817-28.
34.
Moreau JM, Dhariwala MO, Gouirand V, et al. Regulatory T cells promote innate inflammation after skin barrier breach via TGF-beta activation. Sci Immunol 2021; 6: eabg2329.
35.
Salim A, Emerson RM, Dalziel KL. Successful treatment of severe generalized pustular psoriasis with basiliximab (interleukin-2 receptor blocker). Br J Dermatol 2000; 143: 1121-2.
36.
Owen CM, Harrison PV. Successful treatment of severe psoriasis with basiliximab, an interleukin-2 receptor monoclonal antibody. Clin Exp Dermatol 2000; 25: 195-7.
37.
Wallace C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet 2021; 17: e1009440.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.