Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
CLINICAL RESEARCH
Systematic druggable genome-wide Mendelian randomization identifies therapeutic targets for basal cell carcinoma
1
Shandong First Medical University, Jinan, Shandong, China
2
Department of Gastrointestinal Surgery, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, China
3
Department of Burns and Plastic Surgery, Weihai Central Hospital Affiliated to Qingdao University, Weihai, Shandong, China
Submission date: 2025-05-23
Final revision date: 2025-07-30
Acceptance date: 2025-09-05
Online publication date: 2025-11-06
Corresponding author
Benjia Liang
Department of Gastrointestinal Surgery Shandong Provincial Hospital Affiliated to Shandong First Medical University Jinan, Shandong China
Department of Gastrointestinal Surgery Shandong Provincial Hospital Affiliated to Shandong First Medical University Jinan, Shandong China
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Basal cell carcinoma (BCC) is the most common type of skin cancer, with its incidence increasing annually, posing a significant challenge to public health. Currently, the treatment of BCC mainly includes surgical resection, radiotherapy, and pharmacotherapy. However, for high-risk or recurrent BCC cases, traditional treatments may be limited in efficacy, and there is an urgent need to explore more effective targeted therapeutic strategies. This study aims to identify and validate potential druggable genes for BCC treatment by integrating multi-omics and pharmacogenomics approaches.
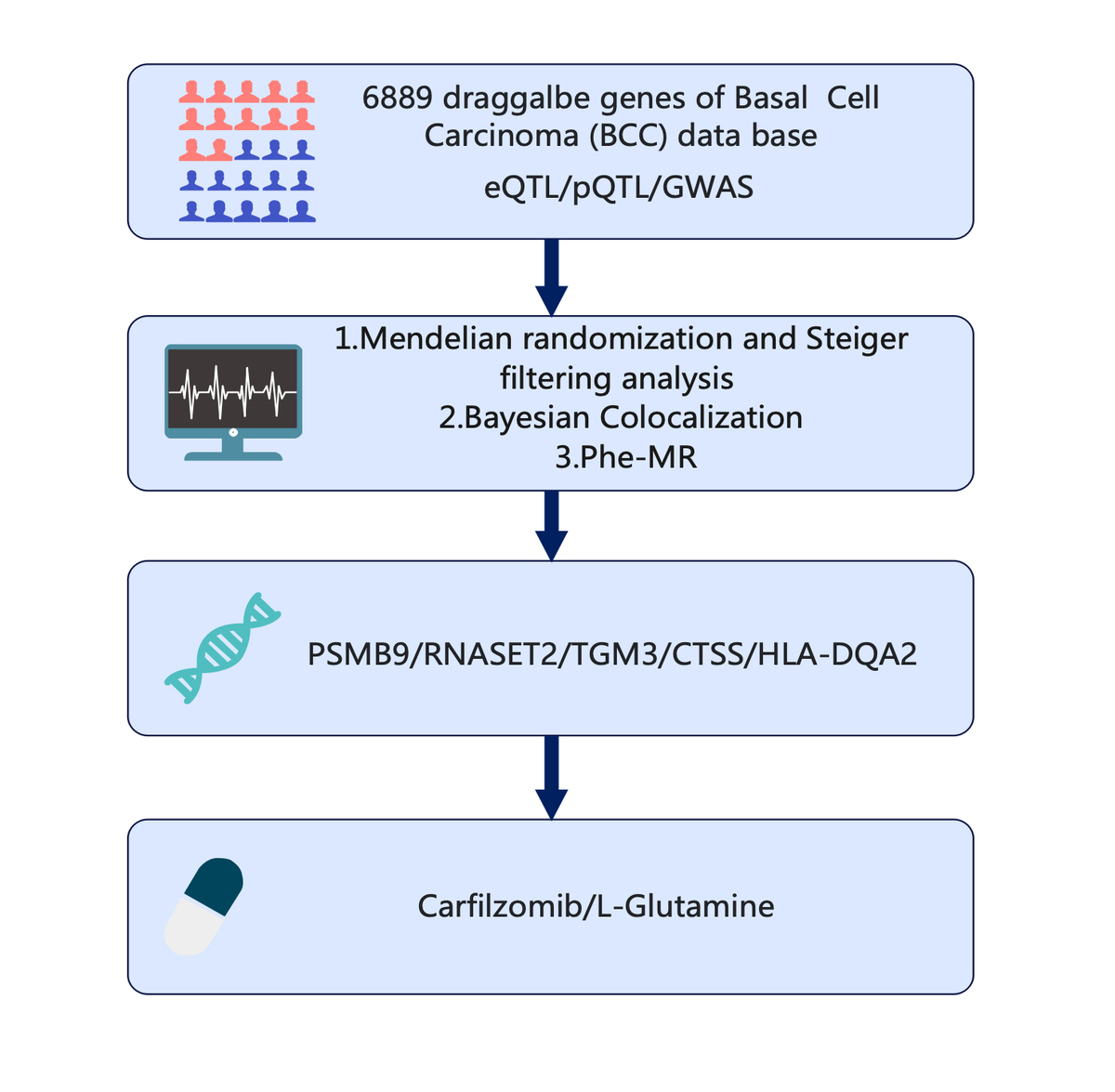
Material and methods:
Utilizing pharmacogenomics, transcriptomics, proteomics, and genome-wide association study (GWAS) data, we employed Mendelian randomization (MR) and Bayesian colocalization analyses to identify genes associated with BCC development. Phenome-wide Mendelian randomization (Phe-MR) analysis was further conducted to elucidate the causal relationships between these genes and various disease phenotypes.
Results:
The study identified PSMB9, TGM3, CTSS, HLA-DQA2, and RNASET2 as potential drug targets, with PSMB9 and RNASET2 positively correlated with BCC risk, while CTSS showed a negative correlation. Additionally, carfilzomib and L-glutamine were identified as existing compounds with potential therapeutic agents.
Conclusions:
The strength of this study lies in its integrative approach, which not only enhances the reliability of the findings but also provides new possibilities for targeted drug development. Phe-MR analysis ensured the safety of the candidate genes and provided guidance for future targeted drug development. The results highlight the importance of further exploring these druggable genes and underscore the value of MR analysis in drug discovery, offering new therapeutic strategies for BCC and directions for future research.
Basal cell carcinoma (BCC) is the most common type of skin cancer, with its incidence increasing annually, posing a significant challenge to public health. Currently, the treatment of BCC mainly includes surgical resection, radiotherapy, and pharmacotherapy. However, for high-risk or recurrent BCC cases, traditional treatments may be limited in efficacy, and there is an urgent need to explore more effective targeted therapeutic strategies. This study aims to identify and validate potential druggable genes for BCC treatment by integrating multi-omics and pharmacogenomics approaches.
Material and methods:
Utilizing pharmacogenomics, transcriptomics, proteomics, and genome-wide association study (GWAS) data, we employed Mendelian randomization (MR) and Bayesian colocalization analyses to identify genes associated with BCC development. Phenome-wide Mendelian randomization (Phe-MR) analysis was further conducted to elucidate the causal relationships between these genes and various disease phenotypes.
Results:
The study identified PSMB9, TGM3, CTSS, HLA-DQA2, and RNASET2 as potential drug targets, with PSMB9 and RNASET2 positively correlated with BCC risk, while CTSS showed a negative correlation. Additionally, carfilzomib and L-glutamine were identified as existing compounds with potential therapeutic agents.
Conclusions:
The strength of this study lies in its integrative approach, which not only enhances the reliability of the findings but also provides new possibilities for targeted drug development. Phe-MR analysis ensured the safety of the candidate genes and provided guidance for future targeted drug development. The results highlight the importance of further exploring these druggable genes and underscore the value of MR analysis in drug discovery, offering new therapeutic strategies for BCC and directions for future research.
REFERENCES (53)
1.
Sethi A, Vogt M, Brocato J, Habeeb L, Squittieri N. Comment on “Basal cell carcinoma: Contemporary approaches to diagnosis, treatment, and prevention”. J Am Acad Dermatol 2023; 88: e29-30.
2.
Sobjanek M, Zabłotna M, Michajłowski I, Nedoszytko B, Lesiak A, Nowicki R. -308 G/A TNF-α gene polymorphism influences the course of basal cell carcinoma in a Polish population. Arch Med Sci 2015; 11: 599-604.
3.
Untaaveesup S, Dendumrongsup W, Srichana P, et al. Clinical outcomes and adverse events of Hedgehog pathway inhibitors for advanced basal cell carcinoma patients: a systematic review and meta-analysis. Heliyon 2025; 11: e39476.
4.
Wang R, Chen Y, Shao X, et al. Burden of skin cancer in older adults from 1990 to 2021 and modelled projection to 2050. JAMA Dermatol 2025; 161: 715-22.
5.
Pan Y, Tang B, Guo Y, Cai Y, Li YY. Global burden of non-melanoma skin cancers among older adults: a comprehensive analysis using machine learning approaches. Sci Rep 2025; 15: 15266.
6.
Zhou L, Zhong Y, Han L, Xie Y, Wan M. Global, regional, and national trends in the burden of melanoma and non-melanoma skin cancer: insights from the global burden of disease study 1990-2021. Sci Rep 2025; 15: 5996.
7.
Si Z, Ying J, Zhou Y. A study on the global burden of non-melanoma skin cancer from 1990 to 2019. Arch Med Sci 2024; 20: 1902-8.
9.
Kilgour JM, Jia JL, Sarin KY. Review of the molecular genetics of basal cell carcinoma; inherited susceptibility, somatic mutations, and targeted therapeutics. Cancers (Basel) 2021; 13: 3870.
10.
Cameron MC, Lee E, Hibler BP, et al. Basal cell carcinoma: epidemiology; pathophysiology; clinical and histological subtypes; and disease associations. J Am Acad Dermatol 2019; 80: 303-17.
11.
Lesiak A, Slowik-Rylska M, Rogowski-Tylman M, Sysa-Jedrzejowska A, Norval M, Narbutt J. Risk factors in Central Poland for the development of superficial and nodular basal cell carcinomas. Arch Med Sci 2010; 6: 270-5.
12.
Miller SJ. Biology of basal cell carcinoma (Part I). J Am Acad Dermatol 1991; 24: 161-75.
13.
Basset-Seguin N, Herms F. Update in the management of basal cell carcinoma. Acta Derm Venereol 2020; 100: adv00140.
14.
Draper E, Li YY, Mahadevan NR, Laga AC, Hanna J, Russell-Goldman E. Clinicopathologic and molecular characterization of basal cell carcinoma arising at sun-protected sites. Am J Surg Pathol 2025; 49: 328-35.
15.
Peris K, Fargnoli MC, Garbe C, et al. Diagnosis and treatment of basal cell carcinoma: European consensus-based interdisciplinary guidelines. Eur J Cancer 2019; 118: 10-34.
16.
Schmults CD, Blitzblau R, Aasi SZ, et al. Basal Cell Skin Cancer, Version 2.2024, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2023; 21: 1181-203.
17.
Danial C, Sarin KY, Oro AE, Chang AL. An investigator-initiated open-label trial of sonidegib in advanced basal cell carcinoma patients resistant to vismodegib. Clin Cancer Res 2016; 22: 1325-9.
18.
Freshour SL, Kiwala S, Cotto KC, et al. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res 2021; 49: D1144-51.
19.
Finan C, Gaulton A, Kruger FA, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med 2017; 9: eaag1166.
20.
Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018; 361: 769-73.
21.
Yin KF, Chen T, Gu XJ, et al. Systematic druggable genome-wide Mendelian randomization identifies therapeutic targets for sarcopenia. J Cachexia Sarcopenia Muscle 2024; 15: 1324-34.
22.
The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020; 369: 1318-30.
23.
Ferkingstad E, Sulem P, Atlason BA, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet 2021; 53: 1712-21.
24.
Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol 2012; 41: 161-76.
25.
Shu M, Yates TB, John C, et al. Providing biological context for GWAS results using eQTL regulatory and co-expression networks in Populus. New Phytol 2024; 244: 603-17.
26.
Sun X, Chen B, Qi Y, et al. Multi-omics Mendelian randomization integrating GWAS, eQTL and pQTL data revealed GSTM4 as a potential drug target for migraine. J Headache Pain 2024; 25: 117.
27.
Carter AR, Gill D, Davies NM, et al. Understanding the consequences of education inequality on cardiovascular disease: mendelian randomisation study. BMJ 2019; 365: l1855.
28.
Yao M, Mei F, Zou K, Li L, Sun X. A Bayesian bias-adjusted random-effects model for synthesizing evidence from randomized controlled trials and nonrandomized studies of interventions. J Evid Based Med 2024; 17: 550-8.
29.
Liang YY, Zhou M, He Y, et al. Observational and genetic evidence disagree on the association between loneliness and risk of multiple diseases. Nat Hum Behav 2024; 8: 2209-21.
30.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015; 44: 512-25.
31.
Andersen JV, Christensen SK, Aldana BI, Nissen JD, Tanila H, Waagepetersen HS. Alterations in cerebral cortical glucose and glutamine metabolism precedes amyloid plaques in the APPswe/PSEN1dE9 mouse model of Alzheimer’s disease. Neurochem Res 2017; 42: 1589-98.
32.
Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018; 50: 693-8.
33.
Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 2017; 13: e1007081.
34.
Wu S, Meena D, Yarmolinsky J, et al. Mendelian randomization and bayesian colocalization analysis implicate glycoprotein VI as a potential drug target for cardioembolic stroke in South Asian populations. J Am Heart Assoc 2024; 13: e035008.
35.
Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014; 10: e1004383.
36.
Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer 2021; 21: 298-312.
37.
Rouette A, Trofimov A, Haberl D, et al. Expression of immunoproteasome genes is regulated by cell-intrinsic and -extrinsic factors in human cancers. Sci Rep 2016; 6: 34019.
38.
Georgoulis V, Haidich AB, Bougioukas KI, Hatzimichael E. Efficacy and safety of carfilzomib for the treatment of multiple myeloma: an overview of systematic reviews. Crit Rev Oncol Hematol 2022; 180: 103842.
39.
Zhang W, Wu C, Zhou K, et al. Clinical and immunological characteristics of TGM3 in pan-cancer: a potential prognostic biomarker. Front Genet 2022; 13: 993438.
40.
Feng Y, Ji D, Huang Y, et al. TGM3 functions as a tumor suppressor by repressing epithelial to mesenchymal transition and the PI3K/AKT signaling pathway in colorectal cancer. Oncol Rep 2020; 43: 864-76.
41.
Hu JW, Yang ZF, Li J, et al. TGM3 promotes epithelial-mesenchymal transition and hepatocellular carcinogenesis and predicts poor prognosis for patients after curative resection. Dig Liver Dis 2020; 52: 668-76.
42.
Smirnov A, Anemona L, Montanaro M, et al. Transglutaminase 3 is expressed in basal cell carcinoma of the skin. Eur J Dermatol 2019; 29: 477-83.
43.
Chermnykh ES, Alpeeva EV, Vorotelyak EA. Transglutaminase 3: the involvement in epithelial differentiation and cancer. Cells 2020; 9: 1996.
44.
Savoia P, Veronese F, Camillo L, Tarantino V, Cremona O, Zavattaro E. Multiple basal cell carcinomas in immunocompetent patients. Cancers (Basel) 2022; 14: 3211.
45.
Cluntun AA, Lukey MJ, Cerione RA, Locasale JW. Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer 2017; 3: 169-80.
46.
Smyth P, Sasiwachirangkul J, Williams R, Scott CJ. Cathepsin S (CTSS) activity in health and disease - a treasure trove of untapped clinical potential. Mol Aspects Med 2022. 88: 101106.
47.
Li Y, Li Q, Cao Z, Wu J. Multicenter proteome-wide Mendelian randomization study identifies causal plasma proteins in melanoma and non-melanoma skin cancers. Commun Biol 2024; 7: 857.
48.
Schaafsma E, Fugle CM, Wang X, Cheng C. Pan-cancer association of HLA gene expression with cancer prognosis and immunotherapy efficacy. Br J Cancer 2021; 125: 422-32.
49.
Aboulaghras S, Piancatelli D, Taghzouti K, et al. Meta-analysis and systematic review of HLA DQ2/DQ8 in adults with celiac disease. Int J Mol Sci 2023; 24: 1188.
50.
Adolphe C, Xue A, Fard AT, Genovesi LA, Yang J, Wainwright BJ. Genetic and functional interaction network analysis reveals global enrichment of regulatory T cell genes influencing basal cell carcinoma susceptibility. Genome Med 2021; 13: 19.
51.
Bruno A, Noonan DM, Valli R, et al. Human RNASET2: a highly pleiotropic and evolutionary conserved tumor suppressor gene involved in the control of ovarian cancer pathogenesis. Int J Mol Sci 2022; 23: 9074.
52.
Liu Y, Zhang Z, Xi P, et al. Systematic analysis of RNASET2 gene as a potential prognostic and immunological biomarker in clear cell renal cell carcinoma. BMC Cancer 2023; 23: 837.
53.
Han QJ, Zhu YP, Sun J, Ding XY, Wang X, Zhang QZ. PTGES2 and RNASET2 identified as novel potential biomarkers and therapeutic targets for basal cell carcinoma: insights from proteome-wide mendelian randomization, colocalization, and MR-PheWAS analyses. Front Pharmacol 2024; 15: 1418560.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.