Introduction
Cervical cancer is the second most common malignancy, leading to a higher mortality rate in women than breast cancer [1, 2]. It is estimated that 529,800 new cases are diagnosed every year worldwide [3]. The current poor prognosis for cervical cancer is mainly based on the following factors: late FIGO stage, large tumor volume, deep interstitial infiltration, vascular tumor thrombus, and lymph node metastasis. However, sometimes these clinicopathological features are not sufficient to accurately assess the risk of recurrence and metastasis of cervical cancer [4, 5]. Although many molecular markers have been shown to be associated with the prognosis of cervical cancer, the detailed mechanisms of these progress remain unclear.
Long non-coding RNA (lncRNA) is a type of non-coding RNA with a sequence length of more than 200 nucleotides, which play various biological functions such as chromatin modification, transcription, translation, splicing, epigenetic regulation, and so on [6]. Studies have confirmed that lncRNA played an important role in the diagnosis and treatment of tumors [7]. A variety of differentially expressed lncRNAs can be used as biomarkers for cancer or as monitor indicators for patients receiving chemotherapy [8]. LncRNA can also be used as a tumor marker. For example, lncRNA DD3/PCA is highly specific in prostate cancer and has been used as a biomarker for prostate cancer [9].
TP73 antisense RNA 1T (TP73-AS1) is an lncRNA transcribed from chromosome 1p36 and is reported to be involved in cell proliferation and tumor progression [10]. It has been found in hepatocellular carcinoma, breast cancer, glioma, esophageal squamous cell carcinoma and ovarian cancer, and promotes cell proliferation and tumorigenesis of these types of cancer [11, 12]. However, the role of TP73-AS1 in cervical cancer is still unclear.
This study proved that lncRNA TP73-AS1 was up-regulated in cervical cancer tissues and cell lines. COX multivariate analysis showed that high expression of TP73-AS1 could be considered as an independent prognostic factor. In addition, lncRNA TP73-AS1 could promote cervical cancer cell proliferation, migration and invasion, and knockdown of TP73-AS1 inhibited the growth of cervical cancer cells in vivo.
Material and methods
Patients and tumor tissue collection
A total of 50 paired cervical cancer tissues and adjacent non-tumor tissues were collected from the patients via resection of hysterectomy at Shanghai Tenth People’s Hospital. The patients were diagnosed via pathology examination and confirmed with imaging modalities for the cervical cancer, and were recruited from Shanghai Tenth People’s Hospital. The clinicopathological characteristics of the patients are shown in Table I. Only 1 patient was lost to follow-up, and the rate of loss to follow-up was 1/50. The present study was approved by Shanghai Tenth People’s Hospital, and all the patients signed written informed consent.
Table I
Correlation between LncRNA TP73-AS1 expression and clinical features (n = 50). Low/high by the sample median. Pearson χ2 test. *p < 0.05 was considered statistically significant. Clinical features include age, tumor size, FIGO stage, differentiation, histology, SCC-AG, lymph node metastasis and depth of cervical invasion
Animal experiments
Four-week-old model mice (BALB/c nude mice) and control normal mice were purchased from the Model Animal Institute of Nanjing University. During the experiment, the mice were kept under constant temperature conditions, and the standard feed and drinking water were freely provided, and the lights alternated for 12 h. All animal experiments were conducted on the basis of the “Guidelines for the Care and Use of Laboratory Animals” [13].
Cell culture
All cell lines were cultured in RPMI in 1640 medium (10% fetal bovine serum), with 5% oxygen carbon, at 37°C in the incubator. The cells were passaged every 3–4 days, and the experiment was performed when the cells were in the logarithmic growth phase.
RNA extraction and qRT-PCR
RNA extraction: The cells were washed 3 times with PBS and then collected by pre-chilled Trizol reagent. Chloroform was added to the lysates and they were centrifuged for 15 min, then an equal volume of isopropanol was added and they were centrifuged for 15 min, and washed with 75% ethanol twice. The RNAs were air dried then RNA concentration was measured by an ultraviolet spectrophotometer and reverse transcription (RT) was performed using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). Quantitative PCR (qPCR) was performed Using an ABI PRISM 7700 System and TaqMan reagents (Applied Biosystems). The relative expression level was calculated using the 2–ΔΔCt method.
MTT experiment
The MTT assay was performed as previously described [14]. Briefly, the HeLa cells and SiHa cells were collected, and the cell concentration was adjusted to 1 × 104/ml, then the cells were seeded into 96-well plates at 100 μl/well. The cells were conventionally cultured overnight and the cells were transfected with shNC and shTP73-AS1. During the culture process, the cell growth was observed at different time points after transfection, 0, 24, 48, 72, 96 h. 20 μl (5 mg/ml) of MTT solution was added to each well at the specified times, and culture was continued in the cell incubator. After incubating for 4 h at 37°C, the supernatant was removed, and then 200 μl of DMSO was added to each well, and the plate was shaken for 10 min to fully dissolve the crystals. The OD value was determined at 450 nm wavelength by the colorimetric microplate reader and the results were recorded.
Cloning formation experiment
The logarithmic growth phase cells were digested with 0.25% trypsin and the cells were suspended in DMEM medium containing 20% fetal bovine serum. Two low-concentration agarose liquids of 1.2% and 0.7% were prepared separately. Briefly, we mixed 1.2% agarose and 2 × DMEM medium (containing 2 × antibiotics and 20% calf serum) in a ratio of 1 : 1, injected 3 ml of the mixture then mixed 0.7% agarose and 2 × DMEM medium in a ratio of 1 : 1, add 0.2 ml of the cell suspension to the tube, and injected 1.2% agarose in the bottom plate, forming a double agar layer. The plates were placed in a 37°C 5% CO2 incubator for 10 to 14 days. The cell colony numbers were observed under an inverted microscope and the colony formation rate was calculated.
Total protein extraction and western blot
The cells were collected and washed twice with pre-cooled PBS, then the cells were collected and put into a 1.5 ml EP tube, centrifuged at 4°C, 800 rpm for 5 min, and the precipitate was retained. The cell lysates were prepared by adding 100 μl of protein lysate (added protease inhibitor cocktail II) into the cells, and then cells were digested on ice for 30 min, shake every 5 min, centrifuge at 4°C, 14,000 rpm for 20 min. The supernatant was collected in a new EP tube. All of the antibodies were purchased from Abcam, USA.
Wound-healing assay
SiHa and HeLa cells were seeded in 6-well plates. Wound gap in the cell monolayer was generated by scratching with plastic pipette tip. The cells were washed with PBS to remove debris or the detached cells, and cultured in RPMI 1640 medium for another 24 h before calculating the wound width. On the back of the 6-well plate, and draw a horizontal line, and each hole passes through 5 pieces. Add cells and conventional culture; the method of transfecting cells is the same as before. The next day measure the horizontal line as far as possible. Wash the cells three times with PBS, add the medium, and continue the culture in the incubator. Take samples at 0 and 24 h and take pictures.
Cell invasion test
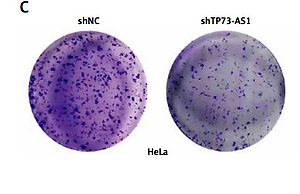
SiHa and HeLa cells were seeded into the upper wells of chamber (Corning, MA, USA) with the Matrigel-coated membrane (BD Biosciences, Franklin Lakes, NJ, USA) in serum-free RPMI 1640. RPMI 1640 with 10% FBS was added to the lower wells. The medium of upper wells and the filters were removed 8 h later. The invasive cells to the bottom of chambers were fixed with 100% methanol and then stained with 0.1% crystal violet 24 h later, imaged and counted under microscope.
Statistical analysis
Statistical analysis was performed using SPSS13.0 software. All the data are expressed as mean ± standard deviation. Survival curves plotted via Kaplan-Meier method and the log-rank test were used to analyze the difference between patients with high or low levels of TP73-AS1 expression and overall survival. The difference among multiple-group (more than two groups) designs was measured with the one-way analysis of variance (ANOVA), and the difference between two groups designs was measured with the Tukey method or Student’s t-test. The overall analysis of the observation data at multiple time points was performed by repeated measures analysis of variance, and multiple comparisons between the two groups or two time points within groups were conducted by the Tukey method or t test, respectively. For co-expressed pairs, Pearson’s correlation coefficient was calculated based on the expression value between every differentially expressed pairs. Multivariate analysis of prognostic parameters in patients with gastric cancer was performed using Cox regression analysis. All p-values are bilateral, and p < 0.05 for the difference has statistical significance.
Results
LncRNA TP73-AS1 was up-regulated in cervical cancer tissues and cell lines and involved in cervical cancer carcinogenesis.
First, qRT-PCR was used to detect the expression of LncRNA TP73-AS1 in 50 cervical cancer samples. The results showed that TP73-AS1 expression was increased in cervical cancer tissues compared to non-tumor tissues (Figure 1 A). In addition, to further investigate the correlation between TP73-AS1 expression and clinical pathological features of cervical cancer patients, the median TP73-AS1 expression in all 50 cervical cancer patients was firstly considered as a cut-off and then the 50 cervical cancer patients were divided into two groups: a high TP73-AS1 expression group (fold change ≥ 2.0, N = 25) and a low TP73-AS1 expression group (N = 25). According to statistical analysis, high expression of lncRNA TP73-AS1 was positively correlated with FIGO stage, lymph node metastasis, and depth of cervical invasion (Figures 1 B–D), and high expression of LncRNA TP73-AS1 was positively related to larger tumor size (p = 0.024) and poor differentiation (p = 0.047), but not related to other clinicopathological features, including age, histology, squamous cell carcinoma antigen (SCC-AG), and lymph node metastasis (Table I, *p > 0.05). Furthermore, Kaplan-Meier analysis (log rank test) showed that cervical cancer patients with high expression of TP73-AS1 had poor overall survival (OS) (Figure 1 E). These results indicated that lncRNA TP73-AS1 was up-regulated in cervical cancer tissues and negatively regulated the overall survival of cervical cancer patients.
Figure 1
LncRNA TP73-AS1 was up-regulated in cervical cancer tissues and cell lines. A–D – QRT-PCR detected the expression level of lncRNA TP73-AS1 in non-tumor tissue and cervical cancer tissue (A), and FIGO stage (B), lymph node metastasis (C), and depth of cervical invasion (D). Student’s t-test: ***p < 0.001. E – Kaplan-Meier method analysis (logrank test) of the overall survival (OS) between lncRNA TP73-AS1 high or low expression cervical cancer patients. F – qRT-PCR was also carried out to detect the expression level of lncRNA TP73-AS1 normal cells Ect1/E6E7 and CC cells including HeLa, SiHa, CASKI, and C33A. Student’s t-test: ***p < 0.001

A further experiment was performed to determine the expression levels of TP73-AS1 in four cervical cancer cells (SiHa, CaSki, HeLa and C33A) and one human cervical immortalized squamous cell (Ect1/E6E7). The qRT-PCR results showed that compared to Ect1/E6E7 cells, the expression of TP73-AS1 was up-regulated in cervical cancer cells (Figure 1 F). COX multivariate analysis showed that high expression of TP73-AS1 could be considered as an independent prognostic factor (Table II), which includes the independent prognostic impact of FIGO stage (*p = 0.046), tumor size (cm) (*p = 0.015), SCC-AG (µg/l) (*p = 0.029) and depth of cervical invasion (**p < 0.001). These results indicated that LncRNA TP73-AS1 was up-regulated in both cervical cancer tissues and cell lines.
Table II
Multivariate analysis of prognostic parameters in patients with gastric cancer by Cox regression analysis. Proportional hazards methods analysis showed the positive, independent prognostic importance of TP73-AS1 expression (**p < 0.001), in addition to the independent prognostic impact of FIGO stage (*p = 0.046), tumor size (cm) (*p = 0.015), SCC-AG (µg/l) (*p = 0.029) and depth of cervical invasion (**p < 0.001). *p < 0.05 was considered statistically significant
LncRNA TP73-AS1 promotes proliferation of cervical cancer cells
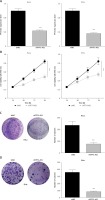
In order to detect the effect of TP73-AS1 on cervical cancer cells, small RNA interference technology was used to interfere with the expression of TP73-AS1 in cervical cancer cell lines including SiHa and HeLa. QRT-PCR results showed that compared with control shNC, the expression of TP73-AS1 was significantly down-regulated in SiHa and HeLa cells transfected with shTP73-AS1 (Figure 2 A). MTT assay showed that cell proliferation ability was significantly decreased in SiHa and HeLa cells transfected with shTP73-AS1 (Figure 2 B). Furthermore, colony formation experiment results showed that the number of cell clones in SiHa and HeLa cells transfected with shTP73-AS1 was significantly lower compared with the control shNC group (Figures 2 C, D). These results suggested that LncRNA TP73-AS1 promoted cervical cancer cell proliferation.
Figure 2
LncRNA TP73-AS1 promotes proliferation of cervical cancer cells. A – QRT-PCR detected the lncRNA TP73-AS1 expression level in HeLa and SiHa cervical cancer cells transfected with shNC and shTP73-AS1 respectively. Student’s t-test: ***p < 0.001. B – MTT assay evaluated cell proliferation ability in shNC or shTP73-AS1 transfected HeLa and SiHa cervical cancer cells. Student’s t-test: ***p < 0.001. C, D – Colony formation experiments detect the number of cell colonies in shNC or shTP73-AS1 transfected HeLa (C) and SiHa (D) cervical cancer cells; the number of cell colonies between shNC and shTP73-AS1 transfected cells was quantified as shown in Figures 2 C and 2 D (right panel). Student’s t-test: ***p < 0.001
LncRNA TP73-AS1 promotes cervical cancer cell migration and invasion
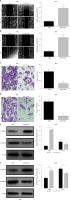
To explore the effect of TP73-AS1 on the migration and invasion abilities of cervical cancer cells, we performed the wound-healing assay and the results showed that the migration rate of cancer cells was significantly decreased in SiHa and HeLa cells transfected with shTP73-AS1 compared with the control shNC (Figures 3 A, B), which indicated that TP73-AS1 promoted the migration rate of cervical cancer cells. Further transwell experiments suggested that when shTP73-AS1 was down-regulated, the invasion rate of cancer cells was also significantly decreased in SiHa and HeLa cells (Figures 3 C, D). Epithelial-mesenchymal transition (EMT) is a pathological process leading to tumor development, which is related to migration and invasion of cancer cells [15]. Cadherin (Cd) conversion, that is, E-cadherin conversion to N-cadherin, is a potential mechanism of tumor progression and migration [16, 17]. The down-regulation of E-cadherin and up-regulation of N-cadherin leads to decreased intercellular adhesion and promotes tumor metastasis migration and invasion, which is an important link in the process of tumor metastasis [18, 19]. Finally, in our study, the expression of E-cadherin conversion to N-cadherin was detected by western blot. The results showed that the expression of E-cadherin was increased, while the expression of N-cadherin was decreased in SiHa and HeLa cells when shTP73-AS1 was down-regulated (Figures 3 E, F). The up-regulated expression of E-cadherin and down-regulated expression of N-cadherin indicated that the migration and invasion of cervical cancer cells were inhibited. Taken together, the above results demonstrated that LncRNA TP73-AS1 promoted the migration and invasion ability of cervical cancer cells.
Figure 3
LncRNA TP73-AS1 promotes cervical cancer cell migration and invasion. A, B – Scratch test determined the migration rate of cells in shNC or shTP73-AS1 transfected HeLa (A) and SiHa (B) cervical cancer cells. The relative wound width (/0h) was measured as Figures 3 A and 3 B (right panel). Student’s t-test: ***p < 0.001. C – Transwell experiments detect the cellular invasiveness in shNC or shTP73-AS1 transfected HeLa (C) cervical cancer cells. D – Transwell experiments detect the cellular invasiveness in shNC or shTP73-AS1 transfected SiHa (D) cervical cancer cells. The invasion cell numbers between shNC and shTP73-AS1 transfected cells were counted as shown in Figures 3 C and 3 D (right panel). Student’s t-test: ***p < 0.001. E, F – Western blot determined the protein accumulation of EMT markers including E-cadherin and D-cadherin. β-actin was used as a loading control. Relative density of western blot bands was quantified by ImageJ (Figures 3 E and 3 F, right panel). Student’s t-test: ***p < 0.001
Knockdown of TP73-AS1 inhibits the growth of cervical cancer cells in vivo
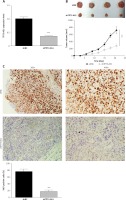
To verify the role of TP73-AS1 in HeLa cell xenograft tumor mice, shNC and shTP73-AS1 were transfected into tumor cells respectively. qRT-PCR results showed that the expression level of TP73-AS1 was significantly decreased in shTP73-AS1 transfected mice (Figure 4 A). The tumor volume was decreased significantly when TP73-AS1 was knocked down (Figure 4 B). Furthermore, immunohistochemistry showed that the Ki67 positive cells were also significantly decreased in TP73-AS1 knockdown mice (Figure 4 C). The results suggested that down-regulation of LncRNA TP73-AS1 suppressed the growth of cervical cancer cells in vivo.
Figure 4
Knockdown of TP73-AS1 inhibits growth of cervical cancer cells in vivo. A – QRT-PCR detected the expression level of lncRNA TP73-AS1 in shNC or shTP73-AS1 transfected mice. Student’s t-test: ***p < 0.001. B – The volume of tumor was compared between normal mouse and LncRNA TP73-AS1 knockdown mouse. The tumor volume was measured at 5 dpi, 10 dpi, 15 dpi, 25 dpi respectively. Student’s t-test: ***p < 0.001. C – Immunohistochemistry experiment detected the Ki67 positive cells in shNC or shTP73-AS1 transfected mice. The Ki67 positive cells were quantified as shown in Figure 3 D (right panel). Student’s t-test: ***p < 0.001
Discussion
This study showed that a high level of TP73-AS1 could be considered as an independent prognostic factor through COX multivariate analysis. In addition, lncRNA TP73-AS1 was up-regulated in cervical cancer tissues and cell lines and promoted the proliferation, migration and invasion of cervical cancer cells, whereas down-regulation of lncRNA TP73-AS1 inhibited the growth of cervical cancer cells in vivo. These findings suggest that lncRNA TP73-AS1 can be used as a new molecular marker for the prognosis of cervical cancer.
FIGO staging late, large tumor volume, deep interstitial infiltration, vascular tumor thrombus and lymph node metastasis are often used as important indicators to evaluate the poor prognosis of cervical cancer, however, these clinicopathological features have certain limitations [20]. Therefore, exploring new molecular markers is necessary for improving the poor prognosis of cervical cancer. This study found that lncRNA TP73-AS1 can be used as a new molecular marker for the prognosis of cervical cancer. At present, many molecular markers related to the prognosis of cervical cancer are found at the protein, DNA and RNA level. Among them, the hypoxia-related proteins, apoptosis-related proteins, cyclooxygenase and epidermal growth factor receptors are common protein markers [21]. Genes are DNA fragments with genetic effects, and genetic abnormalities are inseparable from the development of cervical cancer. General DNA markers are gene copy number alterations (CNAs), chromosomal instability (CIN) and single nucleotide polymorphisms (SNPs) and HPV-DNA [22].
At the RNA level, many studies about microRNAs and some lncRNAs have been reported to reveal their relationship with prognosis of cervical cancer [23]. Studies have shown that miR-214, miR-375, miR-23b, and miR-20 could regulate the motility and invasiveness of cervical cancer cells, and be used as prognostic markers for cervical cancer invasion [24]. Currently, there are many types of cervical cancer-related lncRNAs including HOTAIR, GAS5, LET, and MALATl, which have been shown to be useful as markers for the treatment and prognosis of cervical cancer [25]. Down-regulation of GAS5 can amplify tumor cells by regulating the expression of CDK6, and down-regulation of lncRNA LET can promote tumor metastasis by disrupting the stability of Nuclear factor 90 (NF90), which inhibits tumor cell invasion through hypoxia [26, 27]. In this study, LncRNA TP73-AS1 was up-regulated in cervical cancer tissues and cell lines, which suggested that TP73-AS1 could be considered as a new and independent prognostic factor. A previous study showed that lncRNA TP73-AS1 was significantly decreased in non-small cell lung cancer compared with normal lung tissue. Compared with adenocarcinoma, small cell lung cancer and squamous cell carcinoma, lncRNA TP73-AS1 was significantly up-regulated in large cell lung cancer, suggesting that lncRNA TP73-AS1 may play an important role in the development of tumors [28]. In this study, TP73-AS1 was demonstrated to promote proliferation, migration and invasion of cervical cancer cells.
Ki-67 antibodies are cell proliferation antigens present in the nucleus, which are associated with various types of malignant lesions [29]. In cervical intraepithelial lesions, the expression of Ki-67 was gradually increased with the increase of lesion grade, and is often used as an auxiliary index to evaluate the extent of lesions [30]. In this study, the Ki67 positive cells were significantly decreased in TP73-AS1 knockdown mice, which was consistent with a previous study.
Epithelial-mesenchymal transition (EMT) is thought to be one of the important mechanisms of invasion and migration of carcinoma cells. In the process of EMT, N-cadherin and E-cadherin have the opposite effect, and their expression can promote tumor invasion and migration [31]. E-cadherin plays an important role in maintaining the morphology, structural integrity, and polarity of epithelial cells [32]. This study showed that E-cadherin is an inhibitor of malignant transformation, invasion and migration of tumors, and its down-regulation or loss of function is significantly associated with tumor dedifferentiation, invasive growth, migration and poor prognosis. Loss of E-cadherin is accompanied by up-regulation of N-cadherin expression [33]. This study shows that the expression of N-cadherin is exactly the opposite of that of E-cadherin, which is consistent with previous research. In this study, the up-regulated expression of E-cadherin and down-regulated expression of N-cadherin indicated that the invasion and migration of tumor cells were inhibited, which indirectly suggested that the lncRNA TP73-AS1 may regulate the expression of E-cadherin and N-cadherin to affect the EMT processes, and finally promote the migration and invasion of cervical cancer cells.
Previous studies found that many lncRNAs are differentially expressed in cervical cancer and corresponding adjacent tissues. In this study, we found that lncRNA TP73-AS1 could promote proliferation, migration and invasion of cervical cancer cells and the level of lncRNA TP73-AS1 was upregulated in cervical cancer tissues, which could be considered as a new molecular marker for the prognosis of cervical cancer. It is believed that with the continuous efforts of researchers, more differentially expressed lncRNAs in cervical cancer will be discovered, and the functional mechanism of lncRNA will also be elucidated, which will promote the discovery of molecular markers or therapeutic targets for cervical cancer, and provides more opportunity for the diagnosis and treatment of cervical cancer.