Introduction
Sepsis is a serious life-threatening disease induced by harmful infection [1–4]. Sepsis can lead to multiple complications, and sepsis-induced acute kidney injury (septic AKI) is a commonly observed one [5]. Based on the results of previous findings, septic AKI was found in more than 40% of cases of patients with sepsis, and it can significantly increase the mortality rate of sepsis [6, 7]. Therefore, it is of great importance to further explore the pathogenesis of septic AKI, to find the molecular targets for the diagnosis and treatment of the disease.
Long noncoding RNA (lncRNAs) are a group of non-coding RNAs with the size of over 200 nucleotides [8–10]. In recent years, the roles of lncRNAs in differently diseases have been discussed [11, 12]. Notably, the aberrant expression of lncRNAs has also been found in patients with sepsis-associated AKI, suggesting that lncRNAs may contribute to the pathogenesis of septic AKI [13]. However, the underlying mechanism still requires further investigation.
MALAT1 is a recently identified lncRNA, and the roles of MALAT1 in kidney diseases have been discussed previously [14, 15]. In this study, we will confirm the results with a larger sample size; more importantly, we will explore the underlying mechanism by establishing LPS-induced rat AKI cell models.
Material and methods
Patients
Between May 2015 and July 2019, a total of 15 patients diagnosed with septic AKI at the First Hospital of Jilin University were included in this study, and 15 healthy controls were also included as the control group. Patients with other serious disease, for example, AIDS, cancers, or chronic renal disease, were excluded from this study. The serum samples of the patients and healthy controls were collected and stored at –80°C. Before starting the study, we obtained ethic approval documents from the ethics committee of the First Hospital of Jilin University, and all patients provided the informed consent forms.
Cell culture
Human kidney epithelial cell line HK-2 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Cells were maintained in DMEM medium containing fetal bovine serum (10%, Gibco) in an incubator at 37°C with 5% CO2. When cells reached about 70% confluence, 100 ng/ml LP was added to the cell culture plates to establish the AKI model.
Transfection
MALAT1 siRNA, MALAT1 siRNA NC, MALAT1 over-expression plasmid, miR-23a-3p mimic, miR-23a-3p inhibitor and the negative control were purchased from GenePharma (Shanghai, China). HK-2 cells were transfected by MALAT1 siRNA with or without miR-23a-3p inhibitor using Lipofectamine 3000 (Invitrogen) according to the protocol of the manufacturer.
RT-PCR
To examine the expression of MALAT and miR-23a-3p, the total RNAs were extracted from the HK-2 cells with different treatments by TRIzol (Invitrogen), and reverse transcribed by the Reverse Transcription Kit (Invitrogen). Then polymerase chain reaction (PCR) was performed by SYBR Green methods (the kit was purchased from Invitrogen). U6 was used as the internal for miR-23a-3p and GAPDH was used as the internal control for MALAT. The relative expression of MALAT and miR-23a-3p was quantified by the 2– Δ Δ Ct method.
MTT cell viability assay
Viability of HK-2 cells with different treatments was examined by MTT assay. Briefly, HK-2 cells were placed on 96-well plates and incubated with MTT (500 µg/ml, Beyotime, Shanghai, China) for 4 h. The absorbance at 590 nm was recorded by a microplate reader to determine the viability of the cells.
Cell apoptosis assay
The apoptosis of the HK-2 cells with different treatments was determined by FITC-Annexin V Apoptosis Detection Kit (BD Biosciences, Franklin Lakes, NJ, USA) according to the instructions of the manufacturer using a BD FACSVerse flow cytometer (BD Bioscience) and analyzed by CellQuest software (BD Biosciences).
Western blot analysis
Cells were washed with ice-cold PBS and total proteins were extracted with RIPA lysis buffer. Tissue samples were homogenized in the buffer and subsequently centrifuged to obtain the total protein supernatants. The concentration of total protein was determined using a BCA Protein Assay Kit (Pierce, USA). Equal amounts of protein extracts were separated on 10% sodium dodecyl sulfate polyacrylamide gels (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked in Tris buffered saline Tween 20 (TBST) solution containing 5% skimmed milk at room temperature for 2 h, and then incubated with primary antibodies at 4°C overnight. The protein bands were detected with horseradish peroxidase (HRP)-conjugated secondary antibodies and visualized using an enhanced chemiluminescence detection system (Millipore, USA). Images are shown as representatives of three independent experiments.
Dual-luciferase assay
The target miRNAs of lncRNA MALAT1 were predicted using the starBase online tool. The plasmid containing the 3’-UTR sequences of MALAT paring miR-23a-3p (WT) and the plasmid that included the mutated 3’-UTR sequence of MALAT (MUT) were purchased from GenePharma. The plasmids were transfected into 293T cells by Lipofectamine 3000, with miR-23a-3p mimics or NC (GenePharma). The activity of the luciferase was detected by a commercially available kit (Beyotime) at 48 h after transfection.
RNA pull-down assay
To determine the interaction between lncRNA MALAT1 and miR-23a-3p, an RNA pulldown assay was performed. Briefly, the biotinylated miR-23a-3p probes (purchased from Qingke biotech, Guangzhou, China) were transfected into HK-2 cells. Cells were harvested at 48 h and incubated with Dynabeads M-280 Streptavidin (Solarbio, Beijing, China). Then the bound RNAs were evaluated by RT-qPCR methods.
Statistical analysis
The data are expressed as mean ± standard deviation. The statistical analyses were performed with SPSS software version 22.0 (SPSS, Chicago, IL). Differences between groups were analyzed by analysis of variance or Student’s t-test. P < 0.05 was set as the level of statistical significance.
Results
LncRNA MALAT1 was over-expressed in the serum of septic AKI patients and LPS-treated HK-2 cells
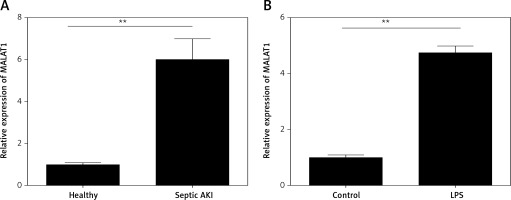
First, the expression levels of MALAT1 in the serum of 15 patients and healthy controls were compared. We found that MALAT1 was dramatically increased in serum of the septic AKI patients when compared with the healthy controls (p < 0.01, Figure 1 A). Moreover, HK-2 cells were treated with LPS to induce the AKI cell model, and we found that MALAT1 was also up-regulated in LPS-treated HK-2 cells in comparison with the control cells (p < 0.01, Figure 1 B).
Figure 1
LncRNA MALAT1 was over-expressed in the serum of the septic AKI patients and LPS-treated HK-2 cells. A – Expression of MALAT1 in septic AKI patients and healthy controls. B – Expression of MALAT1 in LPS-treated HK-2 cells. **P < 0.01
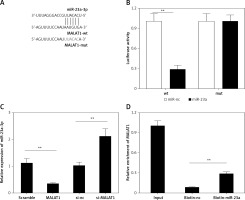
miR-23a-3p is a target of lncRNA MALAT1
Next, using the starBase online bioinformatic tool, miR-23a-3p was predicted as a target miRNA of MALAT1 (Figure 2 A); therefore we further explored whether lncRNA MALAT1 could directly target miR-23a-3p by luciferase reporter assay as well as RNA pull-down assay. As shown in Figure 2 B, the luciferase activity of the WT group was significantly decreased by miR-23a-3p mimics (p < 0.01, Figure 3 B). Also, the expression level of miR-23a-3p was significantly decreased in the MALTA1 over-expression group but markedly increased in the MALTA1 knockdown group (p < 0.01, Figure 3 C). Moreover, results of RNA pull-down analysis showed that the abundance of MALAT1 was significantly higher in the biotin-miR-23a-3p transfected cells than the biotin-NC transfected cells. The above results confirmed that miR-23a-3p is a direct target of lncRNA MALAT1 in HK-2 cells.
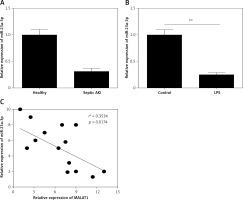
miR-23a-3p was down-regulated in the serum of septic AKI patients and LPS-treated HK-2 cells
Moreover, the expression of miR-23a-3p in the serum of patients and healthy controls was examined. As Figure 3 A shows, miR-23a-3p was significantly decreased in serum of the septic AKI patients (p < 0.01). Moreover, miR-23a-3p was also down-regulated in LPS treated HK-2 cells (p < 0.01, Figure 3 B). Furthermore, results of correlation analysis showed that the expression level of MALTA1 and miR-23a-3p in serum of the septic AKI patients was negatively correlated (p < 0.05, Figure 3 C).
MALAT1 can regulate viability and apoptosis of LPS-treated HK-2 cells via targeting miR-23a-3p
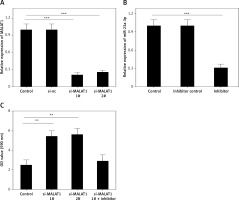
We further explored the effects of MALAT1 on the viability and apoptosis of LPS-treated HK-2 cells. To knock down the expression of MALAT1, we designed two siRNAs, and both markedly decreased the levels of MALAT1 in HK-2 cells (p < 0.01, Figure 4 A). We found that knockdown of MALAT1 significantly increased the viability (p < 0.01, Figure 4 C) and decreased the apoptosis of LPS-treated HK-2 cells (p < 0.01, Figures 4 D, E). Moreover, to explore whether miR-23a-3p was involved in this process, miR-23a-3p inhibitor was added to MALAT1 siRNA transfected HK-2 cells. As Figure 4 B shows, miR-23a-3p inhibitor significantly decreased the level of miR-23a-3p in HK-2 cells (p < 0.01). Furthermore, miR-23a-3p inhibitor partially blocked the proliferative and anti-apoptotic effects of MALAT1 siRNA-treated HK-2 cells (Figures 4 C–E).
Figure 4
MALAT1 can regulate viability and apoptosis of LPS-treated HK-2 cells via targeting miR-23a-3p. A – Effect of MALAT1 siRNA on expression of MALAT1 in HK-2 cells. B – Effect of miR-23a-3p inhibitor on expression of miR-23a-3p in HK-2 cells. C – Results of MTT analysis. D – Results of flow cytometry analysis. E – Quantified results of E. *P < 0.05, **p < 0.01

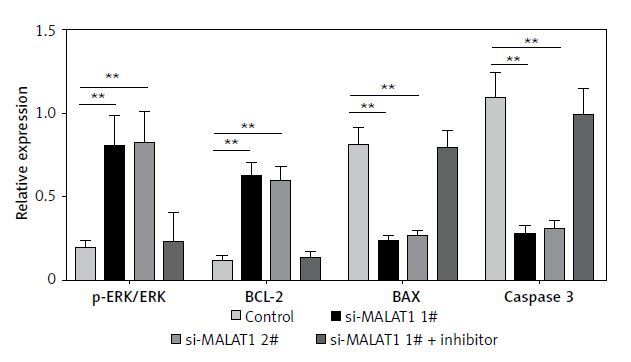
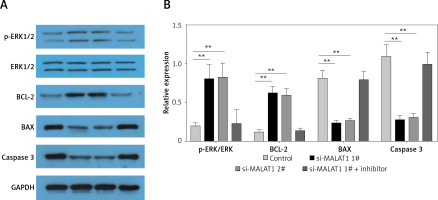
MALAT1 can inhibit ERK signaling in LPS-treated HK-2 cells via targeting miR-23a-3p
Finally, the potential underlying mechanisms of MALAT1 siRNA-induced proliferative and anti-apoptotic effects were explored. We found that MALAT1 siRNA increased the expression of p-ERK and Bcl-2, and decreased the expression of Bax and caspase-3 in LPS-treated HK-2 cells (Figures 5 A, B) and miR-23a-3p inhibitor partially blocked the activation of ERK signaling in MALAT1 siRNA-treated HK-2 cells (Figures 5 A, B).
Discussion
In the present study, we investigated the roles of LncRNA MALAT1 in septic AKI and the possible underlying mechanism. We found that MALAT1 was up-regulated in the serum of septic AKI patients, and using LPS-induced AKI HK-2 cell models we proved that MALAT1 can regulate the proliferation and apoptosis of LPS-treated HK-2 cells via targeting miR-23a-3p through regulating ERK signaling.
The roles of lncRNAs in septic AKI have been discussed in previous studies [14, 15]. However, the roles of MALAT1 in septic AKI are still unclear. In this study, we observed the overexpression of MALAT1 in the serum of septic AKI patients; moreover, we reported for the first time that MALAT1 was over-expressed in LPS-treated HK-2 cells. Taken together, these results suggested that MALAT1 was involved in the pathogenesis of septic AKI, and monitoring the levels of MALAT1 in serum of the patients may be beneficial for the early diagnosis and timely treatment of the disease.
On the other hand, the effects of MALAT1 may not be limited to septic AKI, but may also concern some other kidney diseases. Interestingly, some other reports have discussed the roles of MALAT1 in other kidney disease. For example, downregulation of MALAT1 has been shown to improve the renal function of rats with diabetic nephropathy [14, 16]; moreover, in a very recent publication, MALAT1 was reported to increase the renal fibrosis in chronic kidney disease [17]. Taken together, our results and previous findings have highlighted the roles of MALAT1 in renal diseases as potential therapeutic targets. However, the potential underlying mechanism still requires further investigation.
LncRNAs are known to exert their functions via through binding to the 3′-untranslated regions of the target miRNAs [11, 12, 18–20]. MALAT1 has been reported to interact with different miRNAs and play important roles in the development of different diseases [14, 15]. Using bioinformatic methods, miR-23a-3p has been predicted as a target miRNA of MALAT1, and the results of dual-luciferase reporter assay as well as RNA pull-down analysis confirmed the direct targeting relationship between MALAT1 and miR-23a-3p. Interestingly, miR-23a-3p was down-regulated in serum of the septic AKI patients as well as LPS-treated HK-2 cells, which further confirmed that miR-23a-3p is a direct targeting relationship between MALAT1 and miR-23a-3p in LPS-treated HK-2 cells. Therefore, in clinical application, new therapies designed to inhibit the expression of MALAT1 or increase the expression of miR-23a-3p in septic AKI patients may alleviate the condition of septic AKI via promoting the proliferation and preventing the apoptosis of kidney epithelial cells.
ERK is an important signaling pathway that is involved in the proliferation and apoptosis of cells [21–23]. The roles of ERK signaling in AKI have been discussed previously [24–26]. In this study, knockdown of MALAT1 inhibited activation of ERK signaling in LPS-treated HK-2 cells, and miR-23a-3p inhibitor could reverse the effects. Taken together, these results suggested that MALAT1 can inhibit ERK signaling in LPS-treated HK-2 cells via targeting miR-23a-3p.
The present study has limitations. First, the number of the patients included in this study is relatively small (n = 15), and the results should be evaluated in larger samples; also, the present results should be verified with an in vivo animal study. In future studies, the clinical significance of MALAT1 and miR-23a-3p as indicators of therapeutic efficacy among patients with septic AKI should also be investigated. The expression levels of MALAT1 and miR-23a-3p in serum of the patients before and after the treatment of current first-line medications for sepsis, i.e. vitamin C [27, 28] and steroids [28, 29], should be compared.
In conclusion, our data reveal that MALAT1 can regulate the proliferation and apoptosis of LPS-treated HK-2 cells via targeting miR-23a-3p through regulating ERK signaling. The MALAT1/miR-23a-3p axis could serve as a potential diagnostic biomarker as well as a therapeutic target for the treatment of septic AKI.