Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
HEPATOLOGY / RESEARCH PAPER
Long noncoding RNA profiling and bioinformatics analysis in the human hepatocellular carcinoma cell line HepG2.2.15 treated with different sex hormones
1
The First Affiliated Hospital of Guangdong Pharmaceutical University, China
Submission date: 2020-07-09
Final revision date: 2020-08-28
Acceptance date: 2020-09-05
Online publication date: 2021-05-05
Publication date: 2021-05-05
Corresponding author
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Infection with viral hepatitis remains a significant risk factor for hepatocellular carcinoma (HCC). To understand the long noncoding RNAs (lncRNAs) expression and gender differences in HCC, we measured differentially expressed lncRNAs and mRNAs, aiming to provide new possible diagnosis and prognostic candidates for HCC.
Material and methods:
In our study, we performed lncRNA sequencing to analyze the transcriptome profiles of the human HCC-derived HepG2.2.15 cell line treated with estradiol (E2), testosterone, or untreated. Datasets were analyzed by Coding-Non-Coding Index (CNCI), Coding Potential Calculator (CPC), and other software based on lncRNA levels, such as GO term enrichment analysis and KEGG pathway analysis.
Results:
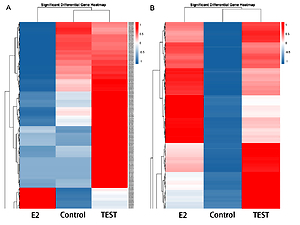
We identified 1203 lncRNAs and 29565 mRNAs that were expressed differentially. A total of 269 lncRNAs and 9429 mRNAs obtained from HepG2.2.15 cells treated with E2 were upregulated, and 552 lncRNAs and 7196 mRNAs were downregulated compared to the control group. A total of 163 lncRNAs and 7185 mRNAs were upregulated and 219 lncRNAs and 5755 mRNAs were decreased in the testosterone group compared to the untreated group. GO analysis demonstrated that the differentially expressed genes were involved in biological processes, such as cellular processes, cell components, and binding. KEGG functional pathway analysis revealed that the top 20 enriched pathways of antisense target genes included dopaminergic synapse, GnRH signaling pathway, FoxO signaling pathway, and MAPK signaling pathway.
Conclusions:
These differentially expressed genes can provide novel insight into the development of more efficient diagnosis and therapeutic strategies for HCC.
Infection with viral hepatitis remains a significant risk factor for hepatocellular carcinoma (HCC). To understand the long noncoding RNAs (lncRNAs) expression and gender differences in HCC, we measured differentially expressed lncRNAs and mRNAs, aiming to provide new possible diagnosis and prognostic candidates for HCC.
Material and methods:
In our study, we performed lncRNA sequencing to analyze the transcriptome profiles of the human HCC-derived HepG2.2.15 cell line treated with estradiol (E2), testosterone, or untreated. Datasets were analyzed by Coding-Non-Coding Index (CNCI), Coding Potential Calculator (CPC), and other software based on lncRNA levels, such as GO term enrichment analysis and KEGG pathway analysis.
Results:
We identified 1203 lncRNAs and 29565 mRNAs that were expressed differentially. A total of 269 lncRNAs and 9429 mRNAs obtained from HepG2.2.15 cells treated with E2 were upregulated, and 552 lncRNAs and 7196 mRNAs were downregulated compared to the control group. A total of 163 lncRNAs and 7185 mRNAs were upregulated and 219 lncRNAs and 5755 mRNAs were decreased in the testosterone group compared to the untreated group. GO analysis demonstrated that the differentially expressed genes were involved in biological processes, such as cellular processes, cell components, and binding. KEGG functional pathway analysis revealed that the top 20 enriched pathways of antisense target genes included dopaminergic synapse, GnRH signaling pathway, FoxO signaling pathway, and MAPK signaling pathway.
Conclusions:
These differentially expressed genes can provide novel insight into the development of more efficient diagnosis and therapeutic strategies for HCC.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.