Introduction
Human glioma is known as the most frequent and primary malignant tumour of the central nervous system (CNS) [1]. Despite various advances in the therapy of glioma, including surgery, radiotherapy, and chemotherapy, patients with glioma remain difficult to treat and have poor prognosis [2]. The terrible prognosis of patients with glioma may be due to the fact that glioma cells with strong invasive ability hijack the normal development of neural progenitor cells to enhance the occurrence of glioma [3, 4]. Therefore, the research began to focus on elucidating the molecular regulatory mechanisms and signalling pathways involved in glioma development and invasion, which may provide an effective theoretical basis for the treatment of glioma.
Runt-related transcription factor 1 (Runx1) has been reported to be a member of the Runx family, which also includes Runx2 and Runx3 [5, 6]. Runx1 is essential for the haematopoiesis with an effect on haematopoietic stem cell generation and regulation [7–9]. Previous studies indicated that Runx1 is involved in human leukaemia with high mutation rate [10, 11]. Recently, Runx1 has been recognised to work in other cancer types including breast cancer [12], prostate cancer [13], endometrial cancer [14], ovarian cancer [15], and non-small-cell lung cancer [16]. Runx1 serves as a downstream direct target of microRNA (miRNA)-139 and exerts its effect on the regulation of malignant behaviour of glioma cells, such as cell proliferation, migration, and invasion [17]. Sangpairoj et al. also revealed that Runx1 could regulate cell migration, invasion, and angiogenesis through the activation of p38 mitogen-activated protein kinase (MAPK) signalling pathway in glioblastoma [18]. However, the mechanism involved in the molecular regulation by Runx1 in glioma is still under investigation.
The Janus kinase and signal transducer and activator of transcription (JAK/STAT) signalling pathway is a highly conserved regulator [19], which mediates the physiological processes by transducing the extracellular signals into regulation of cell growth and differentiation by transcriptional control of target genes [20]. The deregulation of JAK/STAT pathway is involved in many solid tumours, such as breast cancer, prostate cancer, non-small cell lung cancer, glioblastoma, and thyroid cancer [21]. Moreover, it has been reported that Runx1 promotes cancer development by inhibiting SOCS expression and activating of STAT3 signalling pathway in a variety of epithelial cell carcinomas [22]. However, the role of Runx1 and JAK/STAT pathway in glioma remains unclear. Therefore, the suppressor of cytokine signalling 3 (SOCS3) and SOCS4 promoters were selected to study the possible regulatory mechanisms and effects of Runx1 and JAK-STAT signalling pathways on glioma in vivo and in vitro, in order to find further regulatory factors for the occurrence and development of glioma.
Material and methods
Tissue samples
In the present study, 60 patients with glioma were admitted to Guizhou People’s Hospital and enrolled in this study. Then, the tissue was collected from patients who underwent surgical resection without prior chemotherapy or radiotherapy treatment and then stored at –80°C after they were immediately frozen in liquid nitrogen. Clinical stage was determined according to the 2007 WHO CNS tumour classification criteria, including 10 cases of pilocytic astrocytoma of grade I, 11 cases of astrocytomas of grade II, 16 cases of anaplastic astrocytomas of grade III, and 23 cases of glioblastomas of grade IV. According to the grade, the patients were divided into two groups: low-grade group (grade I and II) and high-grade group (grade III and IV). Clinical characteristics were recorded for patients involved in this study (Table I) and all the samples were followed up for 53 months. Informed consent for each patient was obtained prior to the initiation of the study, and the procedures were approved by the Institutional Research Committee (no. 2017GPH028).
Table I
The comparison of clinical characteristics.
Cell culture, plasmid construction, and transient transfection
U-138MG and U-251MG human glioma cell lines, and HEK-293 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Small interfering RNA Runx1 (siRunx1, 5′-ATCACTGGCGCTGCAACAAGAC-3′; 5′-GGATGTT-CCAGATGGCACTCT-3′) were designed and synthesised by Sangon Biotech Co., Ltd (Shanghai, China) and cloned into the pBS/U6 vector (Addgene, Cambridge, MA, USA). A siSTAT3 (5′-GGACAATATCATTGACCTT-3′; 5′-CCACTTTGGTGTTTCATAA-3′) was designed and synthesised by Sangon Biotech Co., Ltd, and cloned into the pBS/U6 vector. Additionally, pcDNA3.1 vector (Addgene) was used for construction of Runx1 expression plasmid. U-138MG and U-251MG cells were transfected with siRunx1, siSTAT3, and their negative control (siNC) via FuGENE Transfection Reagent (Roche Molecular Biochemicals) according to the manufacturer’s instructions.
Real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using the TRIzol® Reagent (Invitrogen) and reversely transcribed with 2 µg of RNA samples by M-MLV Reverse Transcriptase (Invitrogen) to generate complementary DNA. Real-time PCR was carried out using the 7500 Real-Time PCR System (Applied Biosystems) with the Power SYBR Green PCR Master Mix (Takara, Tokyo, Japan) according to the manufacturer’s instructions. Primers of Runx1 used for amplification are listed as following: 5′-TCTTCACAAACCCACCGCAA-3′ (forward) and 5′-GCTCGGAAAAGGACAAGCTC-3′ (reverse). The relative expression of target gene was calculated by 2–ΔΔCt method [23].
Immunohistochemistry (IHC)
Antigen retrieval and immunostaining of Runx1 in paraffin samples were performed as previously described [24]. Sections were heated at 98°C for 15 min in PBS, cooled for 20 min, and added to horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:2,000, Abcam, Cambridge, USA) for 1 h at room temperature. Then, the sealing sections were incubated with rabbit anti-Runx1 monoclonal antibody (No.4334, 1 : 1000, Cell Signaling Technology, Beverly, USA) overnight at 4°C. Chromogenic reaction was performed by adding 3, 3’-diaminobenzidine (DAB) (Solarbio, Beijing, China) and counterstaining with haematoxylin (Solarbio,) for better visualisation.
Western Blot
Total protein (50 µg) isolated from cells were separated on 12% SDS–polyacrylamide gels and transferred to PVDF membranes (Solarbio). The membrane was incubated with the primary antibodies overnight at 4°C after blocking with 5% non-fat milk in Tris-buffered saline (TBS). Antibodies in this study including Runx1 (No.4334), p21 (No.2947), cyclin D1 (No.2978), matrix metalloproteinase-2 (MMP2, No.40994), MMP9 (No.13667), SOCS3 (No.52113), p-STAT3 (No.9145), STAT3 (No.8232), and β-actin (No.4970) were obtained from Cell Signaling Technology (Beverly, USA). SOCS4 (ab170436) antibody was obtained from Abcam. The HRP-conjugated secondary antibodies (ab190492, Abcam) were then detected and quantified by Image J software (NIH Image, Bethesda, USA).
Cell viability assay
MTT assay was performed to detect cell viability. U-138MG and U-251MG human glioma cell lines were seeded onto 96-well plates at a density of 5 × 103 cells/well. Then, 10 µl of 3-(4, 5-dimethlthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide (MTT, 5 mg/ml) was applied and incubated for 4 h at 37ºC. Then the mitochondrial succinate-mediated MTT formazan production was resuspended by adding 100 µl dimethyl sulfoxide (DMSO) to each well. The absorbance of the dissolved formazan was measured at 490 nm by using a scanning microplate spectrophotometer (Bio-Rad, Hercules, USA).
Colony formation assay
For the colony formation assay, U-138MG and U-251MG human glioma cell lines were plated onto 10-cm dishes at a density of 5 × 104 cells per dish. The cells were then cultured for 14 days at 37°C in a 5% CO2 incubator. The number of colonies composed of over 50 cells was counted, and then the colonies were stained for 30 min with 0.5% crystal violet in 20% methanol. Representative images were photographed by camera (Nikon, Tokyo, Japan).
Wound-healing assay
For the wound-healing assay, U-138MG and U-251MG human glioma cells were seeded onto 35-mm dishes until monolayer cells formed. A uniform scratch was then created by using a 10 µl pipette tip. The cells were incubated for another 48 h in a serum-free medium at 37°C in a 5% CO2 incubator. The speed of wound closure could show the ability of cell migration, and representative images were photographed with an inverted microscope (Leica Microsystems, Weitzlar, Germany).
Cell invasion assays
For the cell invasion assay, the transwell chambers were solidified with Matrigel (BD Biosciences, San Jose, USA) by incubating cells at 37°C for 6 h. U-138MG and U-251MG human glioma cells were seeded onto the upper chamber in serum-free DMEM medium with a total number of 4 × 105. Meanwhile, DMEM medium supplemented with 10% FBS was added into the lower chamber, and then invasive cells on the lower chamber were stained after incubation at 37°C for 24 h. Representative images were photographed with an inverted microscope, and cells were counted using the Image J software.
Luciferase reporter assay
The sequences of SOCS3 and SOCS4 promoter were amplified by PCR with human genomic DNA as a template. The fragments were then cloned into the upstream of the luciferase gene in the pGL4.10 basic luciferase vector (Promega, Madison, WI, USA).
HEK-293T cells were transfected with luciferase reporter construct along with Runx1, Runx1-shRNA, or control plasmid. Transfected cells were incubated for 48 h and then lysed for luciferase activity detection by using a dual luciferase assay system (Promega).
Mice and tumour
BALB/c nude mice (4–6 weeks old; Beijing Vital River Laboratory Animal Technology Company, China) were randomly divided into two groups. U-138MG cells (3 × 106) of sh-control or Runx1-shRNA were administered in 100 µl PBS and subcutaneously injected into the dorsal flank of mice in two groups. Tumour volume was calculated once a week using the equation volume (mm3) = (length × width2)/2. The mice were then sacrificed on day 42 post injection. Tumour masses were weighed and tumours were then subjected to IHC and western blot analysis. All animal care and handling procedures were approved by the Institutional Animal Use and Care Committee of Guizhou People’s Hospital (Approval No.2017-S418).
Ethics
All the animal experimental procedures in this study were performed in accordance with the Declaration of Helsinki and conducted according to protocols approved by Guizhou People’s Hospital.
Statistical analysis
All data in this study were shown as the mean ± standard deviation (SD). Results were analysed using either a one-way ANOVA with Bonferroni’s correction for comparison of multiple groups or an independent Student’s t-test for comparison of two groups. The overall survival of mice was shown in a Kaplan-Meier curve. Survival curves were analysed using log-rank test. Data were analysed through GraphPad Prism 6.0 software (San Diego, CA, USA). A value of p < 0.05 was considered to be statistically significant (*p < 0.05, **p < 0.01, or ***p < 0.001).
Results
Runx1 expression is increased in glioma tissues
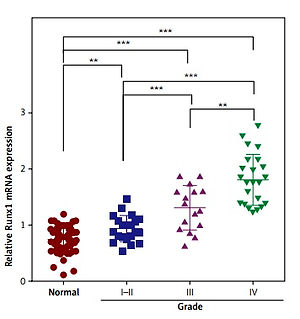
Firstly, the expression levels of Runx1 in human glioma tissues were evaluated by qRT-PCR and immunohistochemistry (IHC). The results of qRT-PCR suggested that Runx1 mRNA expression levels were significantly higher in human glioma tissues than in the adjacent normal tissues. Additionally, increased Runx1 expression in glioma was closely associated with increased tumour grade (Figure 1 A). IHC analysis showed that Runx1 protein expression levels were also significantly increased in glioma tissues compared to the adjacent normal tissues (Figure 1 B). Furthermore, glioma patients in the high Runx1 expression group had remarkably decreased survival rate compared to those in the low Runx1 levels group (Figure 1 C). These data indicated that increased Runx1 expression was implicated in the tumorigenesis of glioma.
Figure 1
Increased expression of Runx1 in the glioma tissues. A – Increased mRNA expression level of Runx1 in the glioma tissues compared to normal tissues assayed (up) and mRNA expression level in glioma tissues with increased tumour grade (down) by qRT-PCR. N = 60, ***p < 0.001 for glioma tissues versus normal tissues. B – Up-regulated Runx1 protein level in the glioma tissues by IHC (200 μm). C – Kaplan-Meier survival curves in glioma patients according to Runx1 expression. High expression: Runx1 levels in the top third; Low expression: Runx1 levels in the bottom third. N = 76, p = 0.0251
Runx1 knockdown inhibits viability and migration of glioma cells
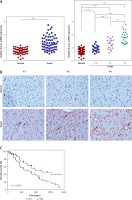
To determine the role of Runx1 in glioma cell viability, overexpression, and knockdown of Runx1 were performed in human glioma cell lines. U-138MG and U-251MG cells were efficiently transfected with Runx1, siRunx1-1#, and siRunx1-2# (Figures 2 A, B). Then, we performed MTT and colony formation assays to evaluate whether Runx1 was involved in glioma cell viability and growth. Our results showed that Runx1 overexpression promoted cell viability and colony formation of U-138MG and U-251MG cells. By contrast, cell viability and colony formation were reduced when Runx1 was knocked down (Figure 2 C, D).
Figure 2
Runx1 knockdown inhibits viability of glioma cells. A – mRNA expression level of Runx1 after transfected with vector, Runx1, siNC, siRunx1-1#, or siRunx1-2# plasmid in the U-138MG and U-251MG glioma cell lines. B – Assessment of Runx1 protein expression level in both of the two cell lines with various treatments. C – MTT assay performed to evaluate cell viability of glioma cells. D – Colony formation assay performed to assess viability of glioma cells
*P < 0.05, **p < 0.01 or ***p < 0.001.
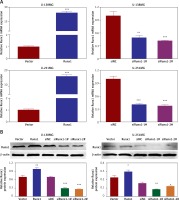

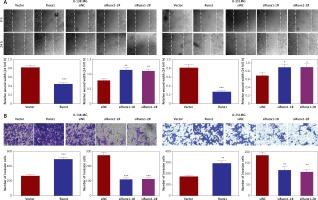
To further evaluate the role of Runx1 in glioma cell migration, a wound-healing assay was used to determine the migration, and transwell assay was conducted to detect the invasion of U-138MG and U-251MG cells. We found that overexpression of Runx1 remarkably increased cell migration and invasion. Downregulation of Runx1 led to the opposite consequence compared to the control cells (Figures 3 A, B).
Figure 3
Runx1 knockdown inhibits migration of glioma cells. A – Wound-healing and migration distances at 0 h and 24 h (100×). B – Cells invading through the transwell chamber (200×) C – Protein expression of p21, cyclinD1, MMP2, and MMP9 in the U-251MG and U-138MG glioma cells transfected with vector, Runx1, siNC, siRunx1-1#, or siRunx1-2# plasmid
*P < 0.05, **p < 0.01, or ***p < 0.001.
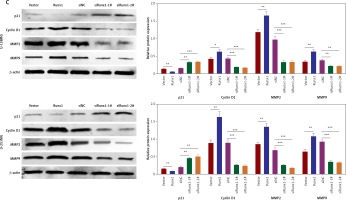
Western blot results showed that overexpression of Runx1 decreased p21 expression and increased cyclin D1 expression. Runx1 knockdown up-regulated the p21 expression and down-regulated cyclin D1 expression in U-138MG and U-251MG cells (Figure 3 C). Previous studies have shown that matrix metalloproteinase-2 (MMP2) and matrix metalloproteinase-9 (MMP9) facilitated tumour invasion and migration by degrading extracellular matrices [25]. We found that MMP2 and MMP9 expression levels increased upon overexpression of Runx1 and decreased as a result of Runx1 knockdown, in both of the two cell lines (Figure 3 C). These results demonstrated that Runx1 knockdown inhibited the development of glioma cells.
Runx1 activates JAK/STAT signalling pathway by inhibiting the activation of SOCS3/SOCS4 promoter
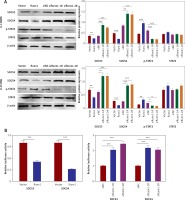
To understand the regulatory mechanism underlying the Runx1 knockdown-induced inhibition of glioma cells development, we examined the protein expression including SOCS3, SOCS4, phosphorylated STAT3 (p-STAT3), and STAT3. We found that overexpression of Runx1 decreased SOCS3/SOCS4 expression and increased p-STAT3 expression in both U-138MG and U-251MG cells. Runx1 knockdown up-regulated the SOCS3/SOCS4 expression and down-regulated p-STAT3 expression in both cell lines (Figure 4 A). Moreover, Figure 4 B showed that Runx1 overexpression significantly decreased the luciferase activity of SOCS3 and SOCS3, while Runx1 knockdown increased the luciferase activity of SOCS3 and SOCS3. Therefore, these results demonstrated that Runx1 inhibited the activation of SOCS3/SOCS4 promoter, and Runx1 contributed to the activation of JAK/STAT signalling pathway.
Figure 4
Runx1 activates JAK/STAT3 signalling pathway and inhibits the activation of SOCS3/SOCS4 promoter. A – Protein expression of SOCS3, SOCS4, p-STAT3, and STAT3 in the U-138MG and U-251MG glioma cells transfected with vector, Runx1, siNC, siRunx1-1#, or siRunx1-2# plasmid. B – Luciferase activity measured after various treatment in the HEK-293T cells
*P < 0.05, **p < 0.01, or ***p < 0.001.
Runx1 promotes viability and migration of glioma cells via JAK/STAT signalling pathway
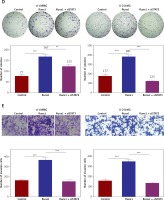
Subsequently, we constructed siSTAT3-1# and siSTAT3-2# plasmids for the STAT3 downregulation, and siSTAT3-2# was chosen in the following experiments because it was more effective at knocking down STAT3 expression (Figures 5 A, B). We examined the effect of downregulation of STAT3 on the viability and migration of glioma cells. Our results indicated that cell viability and colony formation of glioma cells were facilitated in cells transfected with Runx1 plasmid alone, while this effect was reversed owing to co-transfection with siSTAT3 plasmid (Figures 5 C, D). Similarly, the impact of STAT3 knockdown on glioma cell migration and invasion was determined by wound-healing and transwell assay. Glioma cells with Runx1 overexpression showed obviously increased migration and invasive ability. However, STAT3 knockdown reversed this consequence (Figures 5 E, F). Furthermore, Western blot results showed that the increases of p-STAT3, cyclin D1, MMP 2, and MMP 9 and the decrease of p21 induced by Runx1 overexpression were attenuated by STAT3 knockdown (Figure 5 G). Thus, the effect of Runx1 on the promotion of glioma cell viability and migration was abolished when STAT3 was downregulated. Our results suggested that Runx1 promotes viability and migration of glioma cells via JAK-STAT signalling pathway activation.
Figure 5
Runx1 promotes viability and migration of glioma cells via JAK/STAT signalling pathway. (A) mRNA and (B) protein expression level of STAT3 after transfection with siNC, siSTAT3-1#, or siSTAT3-2# plasmid in the U-138MG and U-251MG glioma cell lines. C – MTT analysis of the U-138MG and U-251MG glioma cells transfected with siNC, siSTAT3-1# or siSTAT3-2# plasmid. D – Colony formation assays. E – Images of wound-healing and migration distances at 0 h and 24 h with different treatments F – Cell invasion assays. G – Protein expression of p-STAT3, STAT3, p21, cyclinD1, MMP2, and MMP9 in the glioma cells with different treatments.
*P < 0.05, **p < 0.01, or ***p < 0.001.

Runx1 knockdown suppressed tumour growth in the tumour-bearing mice by inhibition of JAK/STAT signalling pathway
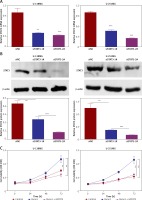
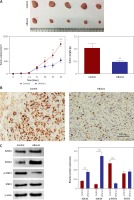
BALB/c nude mice were used to evaluate the effect of Runx1 on tumour progression in vivo. Mice were administrated with U-138MG cells transfected with siRunx1 plasmid or siNC plasmid as a control. The results showed that the tumour volume in the siRunx1 group were significantly smaller than in the control group (Figure 6 A). IHC results suggested that the expression of Ki-67 was decreased in glioma tissues of the siRunx1 group (Figure 6 B). Protein expression of SOCS3/SOCS4 was profoundly increased and p-STAT3 expression was decreased with Runx1 knockdown (Figure 6 C), indicating that Runx1 knockdown inhibited the JAK/STAT signalling pathway. Taken together, these results demonstrate that Runx1 knockdown suppressed tumour growth in vivo by inhibition of JAK/STAT signalling pathway.
Figure 6
Runx1 knockdown suppresses tumour growth in the tumour-bearing mice and inhibits JAK/STAT signal pathway. A – Tumour volume and tumour weight of the siRunx1 group of U-138MG cells and the control group. B – Down-regulated Runx1 protein level in the glioma tissues by IHC. C – Protein expression of SOCS3, SOCS4, p-STAT3, and STAT3 in the U-138MG cells of the siRunx1 group and the control group
*P < 0.05, **p < 0.01, or ***p < 0.001.
Discussion
Gliomas are characterised by high mortality due to the highly proliferative and invasive abilities of glioma cells, even when combining surgery and adjuvant therapies [3, 4, 26]. Studies have focused on the detailed mechanisms underlying this malignant behaviour by examining the role of the brain tumour micro-environment, glioma stem cells, epigenetic control, and dysfunctional signalling pathways such as Wnt pathway and TGF-β pathway [4, 27–29]. In our study, Runx1 was proven to be implicated in the tumourigenesis of human glioma with up-regulated Runx1 in human glioma tissues compared with normal tissues. Runx1 promoted the viability/proliferation and migration of glioma cells by activating the JAK/STAT signalling pathway through targeting of the promoter of SOCS3/SOCS4. Further assays indicated that inhibition of Runx1 suppressed tumour growth in tumour-bearing mice in vivo.
Emerging evidence shows that Runx1 is a very context-dependent gene, which works as both an oncogene and a tumour suppressor gene [30, 31]. Clinical and basic research suggest that loss-of-function mutations owing to the point mutations in the runt domain or chromosomal translocations contribute to the differentiation block and improve malignant behaviours of glioma cells [30, 32]. Runx1 is identified as a tumour suppressor in acute myeloid leukaemia and myelodysplastic syndrome [10, 11]. In contrast, Choi et al. showed that Runx1 is essential for T-cell acute lymphoblastic leukaemia cell survival via activation of oncogenic genes with TAL1 and Notch1 signalling [33]. In addition, Runx1 works as a tumour suppressor in the ER-positive luminal subtype of breast cancer, while exerting its oncogenic effect on triple-negative breast cancers with increased Runx1 expression and poor prognosis [12]. Sangpairoj et al. reported that Runx1 knockdown significantly inhibited the migration and invasion of IL-1β-treated U-87MG human glioblastoma cells [18]. In the present study, our data demonstrated that Runx1 have an oncogenic effect that promotes the viability and migration of glioma cells.
JAK/STAT signalling activation has been shown to be associated with haematological malignancies and several solid tumours [34, 35]. Recent studies demonstrated that STAT3 was also implicated in the tumour progression of glioblastoma [36]. Activation of STAT3 signalling after phosphorylation of Enhancer of Zeste Homologue 2 facilitates self-renewal and tumorigenesis of glioblastoma stem-like cells [37]. Silence of STAT3 suppresses the glioma cell aggression and tumour growth for its role in driving the diffuse infiltration [38]. The JAK2/STAT3 signalling pathway is inhibited by a novel small molecule inhibitor named WP1193, which induces cell-cycle arrest and apoptosis in gliomas to mastem-like cells [39]. In our study, we found that Runx1 inhibited the activation of SOCS3/SOCS4 promoter, which in turn inhibited JAK2 kinase activity, as previously reported. Thus, Runx1 activated the JAK/STAT3 signalling pathway and promoted the viability and migration of glioma cells.
In conclusion, the effects of JAK/STAT signalling and its components on the survival, proliferation, and invasion of cancer cells have been established based on previous studies [20]. Targeting the JAK/STAT signalling pathway might be a promising strategy for drug development and cancer therapy in glioma. In our study, the effect of Runx1-JAK-STAT3 signal on the tumourigenesis of human glioma was verified. Therefore, Runx1 could be a potential candidate to combat human glioma by regulation of JAK/STAT signalling pathway.