Spinal cord injury (SCI) is a traumatic event with a poor prognosis and resulting in early immobility and mortality. However, there is limited knowledge of the mechanisms through which SCI occurs, and a molecular method which could prevent paralysis and the sensory function loss is absent in traditional therapeutic methods [1]. Studies on animal models of SCI have shown that SCI-induced damage consists of initial mechanical damage and secondary injury characterized by neuronal apoptosis in the central nervous system (CNS) leading to expansion of the damage [2]. Neuronal apoptosis is mediated mainly by the B-cell lymphoma-2 (Bcl-2) family proteins including anti-apoptotic and pro-apoptotic members [3]. Identification of mechanisms regulating neuron cell death in the secondary injury will benefit in the management of SCI. A reported tumor suppressor, miR-124, functions in the development of various malignancies such as gastric cancer, cervical cancer, hematological malignancies and hepatocellular carcinoma [4]. It has been confirmed that its SNP, rs531564, was associated with susceptibility of esophageal adenocarcinoma, bladder cancer and gastric cancer [5]. In this study, we hypothesized that rs531564 could be used as an indicator for the prognosis of SCI as it interferes with the singling pathway of miR-124 and BIM. Accordingly, we identified BIM, a protein affecting the apoptosis status of the nervous system in humans, as a direct target of miR-124, and further studied the association between rs531564 and the recovery of SCI.
Methods
A total of 528 patients after acute traumatic SCI were recruited, and peripheral blood samples were obtained from each participant for DNA extraction and genotyping analysis. The research technicians were blind to the participant’s clinical condition. The clinical data were collected systematically from all patients with SCI on admission and discharge. The research staff (unaware of the participants’ information about genotypes) collected data from clinical records and uploaded them into a research database. The study protocol was approved by the institutional ethic research committee and written consent was obtained from each participant.
Total RNA was extracted from the leukocytes in peripheral blood or HT22 cells, a cultured neural cell line, using the mirVana miRNA Isolation Kit (Ambion) according to the manufacturer’s instructions.
The PCR amplification for the quantification of the miR-124 and BIM mRNA was performed using TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and TaqMan Human MiRNA Assay Kit (Applied Biosystems, Foster City, CA, USA).
Cell viability was quantified using Cell Counting kit-8 (CCK-8; Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions.
The full length 3’UTR of the BIM mRNA containing the two miR-124 binding sites was amplified through PCR (forward primer 5’-GCGGTTCTCTTGTGGAGGGG-3’; reverse primer 5’-AGAAGGGGAAACGGCAGACA-3’) and subcloned into the restrictive sites of psi-CHECK-2 vector (Promega, Madison, WI, USA). Site-directed mutagenesis was performed to introduce the mutations of BIM mRNA. HT22 cells were co-transfected with the vectors containing wild-type (WT) or mutant (Mut) 3’-UTR and miR-124 mimics or control (RiboBio) and the luciferase activities were observed with the Dual-Luciferase Reporter Assay System (Promega).
HT22 cell or leukocyte samples were lysed with cell lysis buffer and Western blotting was performed to investigate the level of BIM according to the common routine.
Statistical analysis
Differences in selected variables and Hardy-Weinberg equilibrium were evaluated using the χ2 test and Student’s t test. Differences in the distributions of demographic characteristics and frequencies of genotypes between patients and control subjects were evaluated using Student’s t test (for continuous variables) or Pearson’s χ2 test (for categorical variables). A p-value < 0.05 was considered statistically significant. The statistical analyses above were performed with the SPSS Software version 12.0 (SPSS, Chicago, IL, USA) and were based on two-tailed probability.
Results
Table I presents clinical features by rs531564 genotype. There were no significant differences between women and men by rs531564 genotype (p = 0.708), and different age groups are comparable (p = 0.307). The rates of complete injury on admission for different genotypes were 20.52% and 22.29%. Participants in the CC group has significantly lower acute LOS (length of stay) (p < 0.001) and rehabilitation LOS (p < 0.001).
Table I
Demographic, clinicopathological and genotypic parameters of the participants recruited in this study (LOS, length of stay)
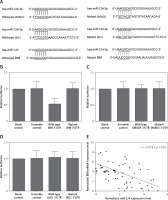
By using online miRNA target prediction tools, we identified BIM, SMAD5 and JAG1 as the candidate target gene of miR-124-3p with the ‘seed sequence’ in the 3’UTR (Figure 1 A). Furthermore, we can see that only the luciferase activity from the cells co-transfected with miR-124-3p and BIM 3’UTR decreased significantly (Figure 1 B), while cells co-transfected with miR-124-3p and SMAD5 or JAG1 3’UTR were comparable (Figures 1 C, D). The results confirmed BIM as a validated target of miR-124-3p in HT22 cells. Also the correlation between the expression level of miR-124 and BIM mRNA showed a negative regulatory relationship (Figure 1 E).
Figure 1
A – BIM, SMAD5 and JAG1 as the candidate target gene of miR-124-3p in HT22 cells with the ‘seed sequence’ in the 3’UTR. B – Luciferase activity reporter assay was conducted to verify BIM as the direct target gene of miR-124. C – Luciferase activity reporter assay verified that SMAD5 was not targeted by miR-124. D – Luciferase activity reporter assay verified that JAG1 was not targeted by miR-124. E – Correlation between expression levels of miR-124 and BIM mRNA (n = 64); they showed a negative regulatory relationship
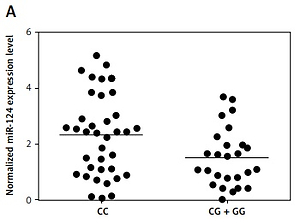
We found that, compared with the CG + GG group, the expression of miR-124-3p was higher in the CC group (Figure 2 A) while the expression of BIM mRNA (Figure 2 B) and protein (Figure 2 C) was lower in the CC group. Moreover, the BIM protein (Figure 2 D) and mRNA expression levels (Figure 2 E) of HT22 cells treated with miR-124a mimics and BIM siRNA were apparently reduced, while the presence of miR-124 inhibitors apparently elevated the BIM protein and mRNA levels, confirming the negative regulatory relationship between miR-124 and BIM.
Figure 2
A – Expression of miR-124 increased in CC group compared with CG + GG group. B – Expression of BIM mRNA decreased in CC group compared with CG + GG group. C – Expression of BIM protein decreased in CC group compared with CG + GG group. D – BIM protein expression level of HT22 cells treated with miR-124a mimics and BIM siRNA was apparently lower than in the scramble control, while in cells treated with miR-124 inhibitors it was apparently higher than in the scramble control. E – BIM mRNA expression level of HT22 cells treated with miR-124a mimics and BIM siRNA was apparently lower than in the scramble control, while in cells treated with miR-124 inhibitors it was apparently higher than in the scramble control. F – Cells transfected with miR-124 inhibitors showed evidently upregulated viability when compared with the scramble controls, while cells transfected with miR-124 mimics and BIM siRNA showed lower viability. G – Number of survival cells transfected with miR-124 mimics and BIM siRNA was increased and the number of apoptotic cells was decreased compared with the scramble controls, while cells transfected with miR-124 inhibitors showed more survival cells and fewer apoptotic cells
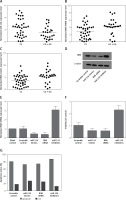
Moreover, cells transfected with miR-124 inhibitors showed evidently upregulated viability (Figure 2 F) when compared with the scramble controls, while cells transfected with miR-124 mimics and BIM siRNA showed comparably lower viability, indicating that miR-124 negatively interfered with the viability of HT22 cells, while BIM positively interfered with the viability of HT22 cells.
As shown in Figure 2 G, compared with the scramble control, the transfection with miR-124 mimics and BIM siRNA elevated the number of survival cells while reducing the number of apoptotic cells. In contrast, cells transfected with miR-124 inhibitors showed fewer survival cells and more apoptotic cells. These results indicated that miR-124 could accelerate proliferation and inhibit apoptosis.
Discussion
The upregulation of BIM has been reported in the pathogenesis of the Parkinson disease (PD) model induced by MPTP [6]. Moreover, it was identified as a target of miR-124 [6]. Further investigations provided evidence that BIM is regulated by other miRNAs in other diseases. For instance, miR-24 could repress the expression of BIM and accordingly lead to suppressed apoptosis in mouse cardiomyocytes [7]. Also miR-363 was shown to inhibit BIM to promote the survival of human glioblastoma stem cells [8]. In our study, we found that miR-124 could inhibit the expression of BIM and accordingly accelerate cell proliferation and inhibit cell apoptosis.
And rs531564, a functional variant in the miR-124-1 gene, known to affect expression levels of the mature miRNA, could lead to allele specific changes in the regulation of the expression of several protein coding genes involved in neuroplasticity and neuro-signaling mechanisms [9] and related to the pathophysiology of abnormal aggression levels [10]. In this study, we found that expression of miR-124 increased in CC-genotyped patients compared with patients carrying the G allele in rs531564. Accordingly, CC genotyped patients not only exhibited reduced BIM mRNA and protein levels, but also suffered from significantly lower acute LOS (length of stay) (p < 0.001) and rehabilitation LOS (p < 0.001).
However, there are limitations to this study. The sample size is relatively small, and a further association study with more participants, especially those with different ethnic backgrounds, is warranted. Also, the regulatory relationship was verified in vivo and in vitro, whereas it was not tested in any animal model. Therefore, a further transgenic animal study is needed to further investigate the role of miR-124 and BIM in the management of SCI.
In conclusion, our research found that down-regulation of miR-124, caused by the presence of rs531564 polymorphism, may increase the expression of BIM, which may promote cell apoptosis and prolong the duration of the recovery stay after SCI. The findings indicated that rs531564 polymorphism could be a predictive biomarker for recovery after SCI.