Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
GENETICS / RESEARCH PAPER
A Multi-Stage Genomic Approach to Uncover Druggable Gene Targets and Neural Pathways in Postpartum Depression
1
The Second Affiliated Hospital of Fujian Medical University
Submission date: 2025-02-24
Final revision date: 2025-05-12
Acceptance date: 2025-06-08
Online publication date: 2025-06-22
Corresponding author
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Background: Postpartum depression (PPD) is a severe emotional disorder affecting women worldwide, with significant impacts on maternal and infant health. Its genetic contributors and biological mechanisms are poorly understood. Identifying druggable genes and clarifying their causal roles may offer insights for more effective treatments.
Material and methods:
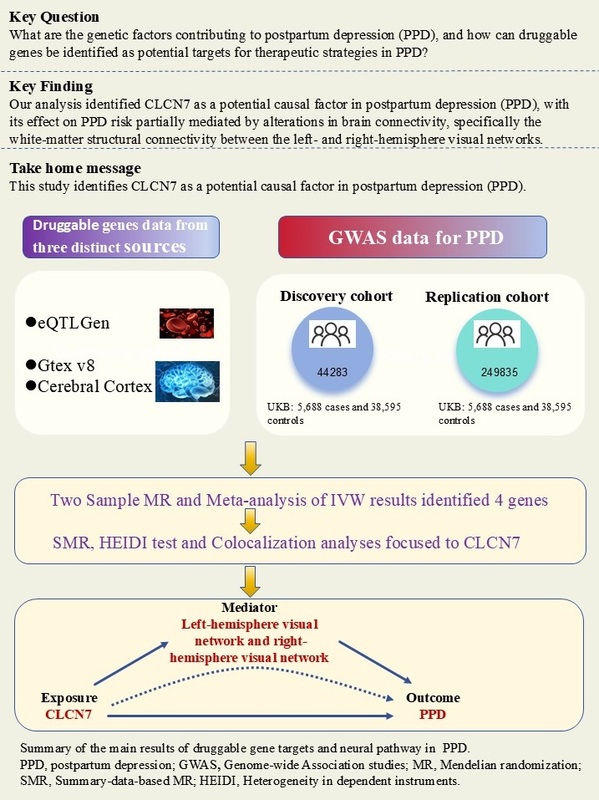
Methods: We identified drug-related genes and screened gene expression quantitative trait loci (eQTL) from the eQTLGen consortium and genotype tissue expression (GTEx) v8 dataset , focusing on 13 brain tissues, along with Qi et al.'s meta-study on the cerebral cortex. Mendelian randomization (MR) analyses were used to investigate causal relationships between gene expression and PPD risk. Replication analyses in an independent PPD cohort validated initial findings, and meta-analysis combined MR results. Summary-data-based MR (SMR) and heterogeneity in dependent instruments (HEIDI) tests were also performed, followed by colocalization analyses to assess shared causal variants. Mediation analyses were conducted to explore how genetic effects may influence brain connectivity patterns.
Results:
Results: From 5,883 druggable genes, 37 were identified with causal links to PPD. Replication confirmed 9 genes, with 4 remaining significant in meta-analysis. SMR and HEIDI analyses focused on CLCN7, which showed robust evidence for causal involvement in PPD. Colocalization analyses suggested shared causal variants, and mediation analyses revealed that CLCN7's genetic risk is partially mediated by left-hemisphere visual network to right-hemisphere visual network white-matter structural connectivity.
Conclusions:
Conclusions: Our analysis identifies CLCN7 as a potential causal factor in PPD, with its effect mediated through brain connectivity. These findings offer targets for future studies and therapeutic strategies for PPD.
Background: Postpartum depression (PPD) is a severe emotional disorder affecting women worldwide, with significant impacts on maternal and infant health. Its genetic contributors and biological mechanisms are poorly understood. Identifying druggable genes and clarifying their causal roles may offer insights for more effective treatments.
Material and methods:
Methods: We identified drug-related genes and screened gene expression quantitative trait loci (eQTL) from the eQTLGen consortium and genotype tissue expression (GTEx) v8 dataset , focusing on 13 brain tissues, along with Qi et al.'s meta-study on the cerebral cortex. Mendelian randomization (MR) analyses were used to investigate causal relationships between gene expression and PPD risk. Replication analyses in an independent PPD cohort validated initial findings, and meta-analysis combined MR results. Summary-data-based MR (SMR) and heterogeneity in dependent instruments (HEIDI) tests were also performed, followed by colocalization analyses to assess shared causal variants. Mediation analyses were conducted to explore how genetic effects may influence brain connectivity patterns.
Results:
Results: From 5,883 druggable genes, 37 were identified with causal links to PPD. Replication confirmed 9 genes, with 4 remaining significant in meta-analysis. SMR and HEIDI analyses focused on CLCN7, which showed robust evidence for causal involvement in PPD. Colocalization analyses suggested shared causal variants, and mediation analyses revealed that CLCN7's genetic risk is partially mediated by left-hemisphere visual network to right-hemisphere visual network white-matter structural connectivity.
Conclusions:
Conclusions: Our analysis identifies CLCN7 as a potential causal factor in PPD, with its effect mediated through brain connectivity. These findings offer targets for future studies and therapeutic strategies for PPD.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.