Introduction
Insomnia is a common sleep disorder characterized by difficulties falling asleep or maintaining sufficient sleep duration, dreaming excessively, waking up easily and returning to sleep with difficulty, or even staying awake throughout the night [1]. Statistical data indicate that the incidence of insomnia among adults in China reaches as high as 38.2%, affecting over 300 million Chinese individuals with sleep disorders [2]. Chronic insomnia poses a significant burden on patients [3]. Although hypnotic medications can temporarily alleviate insomnia symptoms, they may lead to over-reliance on such drugs without addressing the underlying issue [4]. The pathophysiological mechanisms of insomnia are complex and influenced by factors such as sleep environment [5], dietary habits [6], endocrine status [7], psychological issues [8], and circadian rhythms [9]. Increasing evidence suggests that sleep may be related to gut microbiota (GM) [10]. GM are crucial for maintaining bodily homeostasis, including nutrient absorption, metabolism, and toxin degradation. Emerging research suggests a potential bidirectional relationship between sleep regulation and GM functionality through the “microbiome-gut-brain axis” [11]. Clinical studies have confirmed that GM diversity is positively correlated with sleep efficiency and total sleep time, and negatively correlated with wake after sleep onset (WASO) [12]. Animal experimental research has shown that sleep restriction and increased WASO can lead to decreased microbial richness and diversity [13]. The mechanism of the “microbiome-gut-brain axis” between GM and the nervous system reveals the complex interactions between the gut and the brain [14]. GM dysregulation not only affects the metabolic system but may also impact brain function, leading to insomnia, through immune regulation [15], neuroendocrinology [10], and other pathways.
Relevant studies have indicated that individuals with gastrointestinal diseases are more prone to insomnia than the general population [16]. However, identifying specific bacterial species that affect the onset of insomnia among the complex GM remains a research hotspot and challenge. Mendelian randomization (MR), as an advanced causal inference method, uses single-nucleotide polymorphisms (SNPs) as instrumental variables (IVs) for exposure to explore the causal relationship between exposure factors and outcome events [17]. It minimizes biases caused by confounding factors and reverse causality, thereby avoiding reverse causality and common errors in various epidemiological studies. Therefore, this study employed MR analysis to explore the potential causal relationship between GM and insomnia and conducted functional enrichment analysis on the genes adjacent to the IVs to investigate the signaling pathways through which related GM may mediate the occurrence of insomnia. Finally, combining the CTD and Coremine databases, we predict traditional Chinese medicines (TCMs) with potential regulatory effects on the genes adjacent to the IVs, aiming to provide a potential theoretical basis for the integrated traditional Chinese and Western medicine treatment of insomnia.
Material and methods
Study design
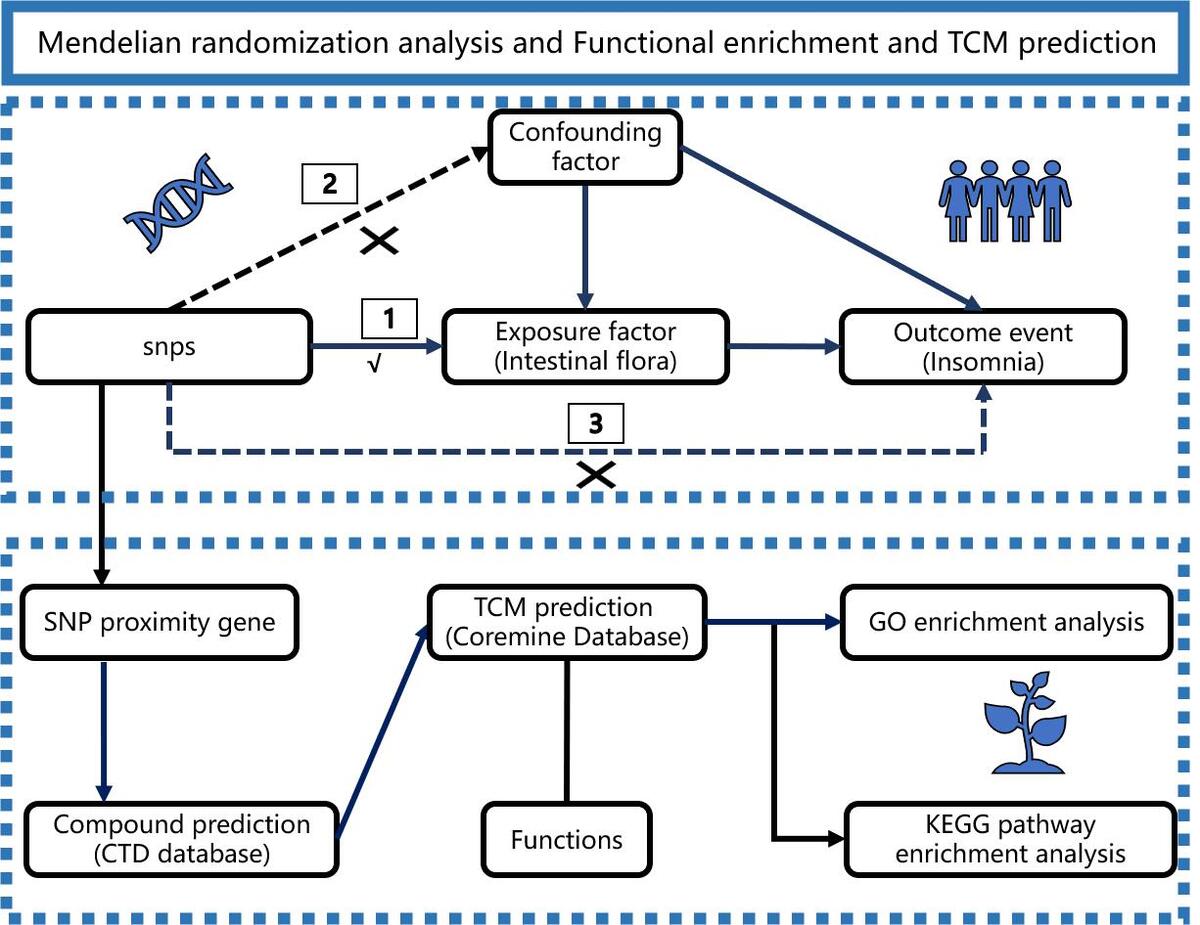
A two-sample MR study was designed to estimate the potential causal link between GM and insomnia. The SNPs were selected as IVs according to three essential premises as follows [18]: (1) SNPs should be strongly associated with GM as the exposure; (2) SNPs should not be associated with confounding factors; and (3) SNPs should not be associated with insomnia as the outcome directly. Subsequently, the adjacent genes of the instrumental variables were obtained, and functional enrichment analysis was conducted to identify the key biological pathways through which the GM mediate the occurrence of insomnia. Furthermore, potential TCMs that regulate this pathway were predicted (Figure 1).
GWAS summary statistics
The GWAS data for GM were sourced from the IEU OpenGWAS database (https://gwas.mrcieu.ac.uk/). Utilizing the ao function of the TwoSampleMR software package in R, we extracted 418 GWAS summary datasets for 211 GM traits from the IEU OpenGWAS database. Similarly, the insomnia data were also obtained from the IEU OpenGWAS database, encompassing 1,402 patients and 485,225 healthy subjects (controls). Both GWAS datasets included populations of European descent, sharing the same genetic background, which allowed for MR studies to be conducted.
Ethical approval
All summary-level datasets in our study were obtained from de-identified public data/studies. Ethical approval and informed consent were previously obtained from the ethics committee. Thus, the requirement for ethical approval was waived for this study.
SNP selection
Firstly, we conducted a screening process to identify SNPs that were highly correlated with exposure at a genome-wide significance level (p < 5 × 10–8). Secondly, we implemented a criterion (r2 < 0.001, kb = 10000) to choose SNPs that were free from dependence on linkage disequilibrium (LD). Thirdly, we excluded SNPs that were not present in the insomnia dataset and palindromic SNPs which have the potential to introduce bias. All of the SNPs for instrumental variables were uploaded to PhenoScanner to identify confounding SNPs associated with insomnia. Based on the assumption of the MR analysis, SNPs used as instrumental variables should be strongly associated with exposure. Subsequently, we ensured the harmonization of exposure and outcome data, confirming that the effect of the SNP on the exposure corresponded to the same allele as its effect on the outcome. Following this, we assessed the possibility of weak instrumental bias by calculating F-statistics, and excluded SNPs with F-statistics less than 10. The F statistic was calculated using the formula F = β2/SE2. Finally, we employed the MR-PRESSO method to identify outlier SNPs. After removing the outliers, the remaining SNPs were used for subsequent MR analysis.
Two-sample Mendelian analysis
Three popular MR methods were employed to assess causal effects: inverse variance weighted (IVW), weighted median and MR-Egger [19, 20]. IVW, a reliable and robust MR method in the absence of horizontal pleiotropy [21], combines the Wald estimates of individual SNPs to derive overall estimates of the effect of GM on insomnia risk. Consequently, the IVW method is broadly acknowledged as the most effective approach to assess causality. Odds ratios (ORs) were utilized to express the effects of GM on insomnia risk. If the result of the IVW method is significant (p < 0.05), it can be considered positive even if other methods yield nonsignificant results, provided that the ORs of those methods line up in the identical direction without heterogeneity or pleiotropy. Two types of IVW approaches, namely the fixed and random effect model, were employed to account for existing heterogeneity. Cochran’s Q test was used to assess the heterogeneity in the IVW method and MR-Egger regression, with a p-value < 0.05 considered statistically significant [22]. Unlike IVW, the MR-Egger method includes an intercept term designed to test for horizontal pleiotropy. A non-zero intercept term indicates that not all genetic variants are valid instruments, thereby biasing IVW estimates. When the instrument strength independent of direct effect (InSIDE) assumption is met, the MR–Egger method can offer an approximation of the causal impact of horizontal pleiotropy. The weighted median method offers a robust effect estimate, even in the presence of unbalanced horizontal pleiotropy (e.g., when 50% of instrumental SNPs are invalid) [23]. Finally, the MR-PRESSO method encompasses three detection functions [24]: horizontal pleiotropic detection, horizontal pleiotropic correction (after outlier removal), as well as assessment of differences in the results of causality estimation before and after correction.
Statistical analysis
Heterogeneity was assessed by employing Cochran’s Q test, where a p-value > 0.05 indicated the absence of heterogeneity. The MR-Egger regression test was utilized to identify horizontal pleiotropy, where a zero-intercept suggests the absence of pleiotropy (p > 0.05).
Reverse MR analysis
To explore the potential causal relationship between insomnia and GM, a reverse MR analysis was carried out, wherein insomnia served as the exposure and GM as the outcome, employing SNPs associated with insomnia as IVs.
All statistical analyses were conducted using R software (version 4.2.3) with the “TwoSampleMR” (version 0.5.6), “MRPRESSO” (version 1.0), and “MendelianRandomization” (version 0.7.0) packages.
Function enrichment of genes linked to instrumental variables and traditional Chinese medicine prediction
Firstly, using the RStudio software package, we identified the proximal genes of the instrumental variables based on the SNP identification numbers, as well as their respective chromosomal sequences and loci. Subsequently, we utilized the CTD database (https://ctdbase.org/) [25] to search for the chemical components corresponding to these proximal genes. By reviewing the literature, we eliminated chemical components with lower support numbers. Then, using Coremine data (https://coremine.com/medical/) [26], we identified TCM that were significantly associated with the aforementioned chemical components, with a screening threshold of p < 0.05. Finally, we employed the David tool (https://david.ncifcrf.gov) to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) [27] enrichment analyses on the proximal genes of the instrumental variables. All enriched pathways and functional terms were filtered using an FDR-adjusted p < 0.05 to ensure biological relevance.
Results
SNP screen results
Based on the established screening criteria, we identified a total of 2,559 SNPs significantly and independently associated with GM. All SNPs exhibited an F-statistic > 10, indicating the absence of weak instrument bias. SNPs associated with the outcome were excluded using the PhenoScanner database (http://www.phenoscanner.medschl.cam.ac.uk/), thereby removing confounded SNPs. Finally, MR-PRESSO was employed to detect outliers and correct for horizontal pleiotropy. Where horizontal pleiotropy was detected among the instrumental variables, outliers were subsequently removed.
MR analysis results
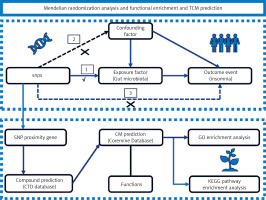
The results of the MR analysis, primarily using the IVW method, showed that the odds ratios (ORs) for Ruminococcaceae (IVW: OR = 1.578; 95% CI: 1.074–2.317; p = 0.020) and Marvinbryantia (IVW: OR = 1.537; 95% CI: 1.062–2.225; p = 0.023) were both greater than 1, indicating that they increase the risk of insomnia. Conversely, the ORs for Pasteurellaceae (IVW: OR = 0.764; 95% CI: 0.599–0.975; p = 0.030), Olsenella (IVW: OR = 0.781; 95% CI: 0.641–0.951; p = 0.014), the Ruminococcus gnavus group (IVW: OR = 0.746; 95% CI: 0.588–0.946; p = 0.016), Mollicutes RF9 (IVW: OR = 0.706; 95% CI: 0.525–0.949; p = 0.021), and Pasteurellales (IVW: OR = 0.764; 95% CI: 0.599–0.975; p = 0.030) were all less than 1, suggesting that they are protective factors against insomnia, reducing the risk of its development. Table I presents the detailed results of the MR analysis. The forest plot (Figure 2) and Circos plot (Figure 3) display the MR analysis results, listing the positive result data in detail.
Table I
Positive results of MR analysis of GM and insomnia
Sensitivity analysis
Both the Cochran’s Q test for IVW and MR-Egger regression indicated no heterogeneity among the SNPs (Table II). The intercept of the MR-Egger regression was nearly zero, suggesting the absence of horizontal pleiotropy. The funnel plot indicated minimal influence of potential bias on the causal relationship. Results from the “leave-one-out” analysis showed that after sequentially excluding each SNP, the IVW analysis results of the remaining SNPs were similar to those obtained when all SNPs were included, with no SNPs found to have a significant impact on the causal association, indicating robustness of the results (Figures 4, 5).
Table II
Quality control results of GM showing a causal relationship with insomnia
Statistical power calculation
In this study, statistical power was calculated as 0.91 by specifying parameters including sample size, Type I error rate (α), case proportion (K), variance explained by the instrumental variables (R2), and odds ratio (OR). This value significantly exceeds the conventional threshold of 80% required for adequate power in typical studies. The analysis demonstrates that sufficient power to reliably identify disease-associated genetic variants was maintained in this study, even under imbalanced case-control ratios.
Functional enrichment analysis of instrumental variable-adjacent genes and prediction of potential traditional Chinese medicines
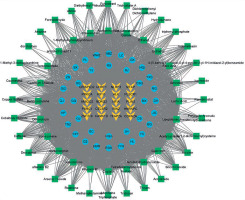
Based on the SNP numbers, their respective chromosomal sequences, and loci, 166 genes corresponding to 84 SNPs were identified. These genes were submitted to the CTD database, yielding 111 chemical compounds represented by lipopolysaccharide, quercetin, and others. Subsequently, 336 TCM were obtained from the Coremine database, which were significantly associated with 80 of these chemical compounds. Cytoscape was used to visualize the top-ranked nodes in terms of degree value within the “gene-chemical component-TCM” mapping network, as shown in Figure 6. Representative TCM with high mapping frequencies include Camellia sinensis root, Ginseng, Radix Curcumae, Salviae Miltiorrhizae Radix, Dried Ginger, Glycyrrhizae Radix and Rhizoma, Aucklandiae Radix, Magnoliae Officinalis Cortex, Scutellariae Radix, Ganodermae Lucidum, and Poria. Finally, functional enrichment analysis was conducted on the genes that have mapping associations with the predicted TCM and chemical components. GO enrichment analysis revealed that these genes are primarily enriched in biological processes such as phosphorylation, mRNA splicing, and hippocampal development; cellular components such as cytoplasm, protein-containing complexes, spliceosomal complexes, and endocytic vesicles; and molecular functions such as protein binding, ATP binding, and protein serine/threonine kinase activity, as shown in Figure 7 A. KEGG pathway enrichment analysis indicated that these genes are mainly enriched in pathways such as neuroactive ligand-receptor interaction and mTOR signaling, as shown in Figure 7 B.
Discussion
The “microbiota-gut-brain axis” represents a complex bidirectional communication system, primarily composed of neural, immune, metabolic, and endocrine pathways, which tightly links the gut and the brain. Abnormal activation of the hypothalamic-pituitary-adrenal (HPA) axis leads to increased cortisol release. Cortisol interacts with immune cells and controls the release of cytokines. Cortisol also disrupts the GM balance, damages the intestinal barrier, and increases proinflammatory cytokines that interfere with sleep, potentially leading to insomnia. The vagus nerve, a crucial nerve for gut-brain communication, can receive and transmit various signals from the gut, such as signals from intestinal immune cells, bacterial metabolites (such as short-chain fatty acids), and neurotransmitters (such as 5-hydroxytryptamine), ultimately affecting brain function and sleep [28].
Numerous studies have indicated a close relationship between the GM and the occurrence and development of insomnia [29]. Recent Mendelian randomization analysis studies have suggested that GM may influence sleep through various metabolic pathways [30]. Therefore, regulating the GM may be an effective therapeutic strategy for improving insomnia symptoms, but the specific GM that affect insomnia remain unclear. Grosicki et al. found that Blautia and Ruminococcus abundance and α-diversity were high, while Prevotella abundance was low, in individuals with better sleep quality [31]. Additionally, some animal experimental results are inconsistent. For example, in mice subjected to fragmented sleep, an increase in Firmicutes and a decrease in Bacteroidetes were observed [32]. In contrast, Maki et al. found that, during chronic sleep deprivation, Bacteroidetes showed a decrease relative to the control group, with reduced Firmicutes abundance, F:B ratio, and α-diversity [33]. In these observational studies, the association between the GM and insomnia is susceptible to confounding factors such as age, environment, dietary patterns, and lifestyle, limiting the establishment of a clear causal relationship between the GM and insomnia. In such cases, MR emerges as a novel method to explore the causal relationship between the GM and insomnia. Based on GWAS summary data for the GM and insomnia, our MR analysis identified seven GM, represented by Ruminococcaceae and the Ruminococcus gnavus group, that have significant genetic causal associations with insomnia. Ruminococcus has been shown to increase in abundance in individuals with higher sleep quality, aligning with our finding that the Ruminococcus gnavus group is associated with a reduced risk of insomnia [31]. Ruminococcaceae can improve sleep quality in insomnia patients (negatively correlated with PSQI and ISI scores) [34] and has beneficial effects on intestinal barrier function, being lower in mice with circadian rhythm disorders [35]. The results of our study, which found an increased risk of insomnia associated with Ruminococcaceae, contrast with some observational studies. Possible reasons include the vulnerability of observational study results to confounding factors, which may introduce bias due to confounding or reverse causality. Our study provides new insights and support for the impact of the GM on insomnia.
TCM applies the holistic view and syndrome differentiation and treatment concepts in clinical practice, leveraging the multi-target and multi-pathway effects of Chinese materia medica, offering unique advantages in preventing and treating insomnia [36, 37]. Many Chinese medicine compound prescriptions, herb pairs and single Chinese herb are claimed to have the effect of “tranquillizing the spirit” and “stabilizing the mind”, thus having a potential clinical effect in treating insomnia [38]. Basic evidence showed that TCM could improve the sleep quality of insomnia model animals. Its mechanism is related to the improvement of GM disorder [39]. This study used the CTD and Coremine databases to further predict, based on the genes adjacent to SNPs, potential TCMs that may intervene in relevant signaling pathways and thereby affect the GM to treat insomnia, mainly including ginseng, Poria cocos, dried ginger, licorice, magnolia bark, Scutellaria root, and Ganoderma lucidum. Ginsenoside Rb1 (Rb1) of ginseng exerts neuroprotective effects through regulation of the abundance of Lactobacillus helveticus [40]. Studies have shown that saponin compounds in ginseng can significantly improve sleep quality indices, shorten sleep latency, and extend sleep duration in mice [41]. Poria cocos has a sweet and mild taste, entering the heart, spleen, and kidney meridians. It strengthens the spleen, removes dampness, calms the mind, and treats insomnia caused by spleen and stomach qi deficiency and damp-phlegm obstruction. The water-soluble polysaccharide, which is the main component of Poria cocos decoction, could significantly improve species richness and diversity in the intestinal flora of rats with chronic sleep deprivation [42]. Studies have found that chemical components such as α-pinene in compound essential oils for calming the mind have sedative and hypnotic effects, reducing sleep latency and extending sleep duration [43]. The orexin system-mediated mTOR signaling pathway is an important part of the downstream signaling network of orexin [44]. Inhibiting mTOR can reduce orexin overexpression, thereby alleviating insomnia episodes, aligning with our study findings.
Domestic and international studies suggest that GM play a significant role in the occurrence and development of insomnia. However, observational study results are susceptible to confounding factors. Given the unique advantages of MR in inferring causal effects, our study innovatively employed MR analysis to explore GM with significant causal associations with insomnia, minimizing the possibility of bias due to residual factors, confounding factors, or reverse causality. Several sensitivity analyses were conducted to satisfy the core assumptions of MR, ultimately identifying genetic variants highly and independently correlated with the phenotype, excluding those associated with potential confounding factors, and ensuring the accuracy of the results. MR studies are affected by pleiotropy, and our study used MR-Egger regression to ensure robustness. Based on this, predicting potential interventional Chinese materia medica through MR instrumental variable adjacent genes is of great significance in the prevention and treatment of insomnia, providing a reference for subsequent research on new anti-insomnia Chinese materia medica. This study has some limitations: 1. All GWAS data selected were from European populations, so the applicability of our conclusions to other populations needs further verification. 2. Single genetic factors cannot explain all phenotypic variations, and our study could not consider environmental factors. 3. As MR analysis is a research method based on genetic inference of causality, it can only provide potential causal relationships and cannot determine the specific biological pathways leading to these causal relationships. Future studies can use a combination of bioinformatics analysis and experiments to explain and validate potential molecular mechanisms.
In conclusion, in this study, we found that the MR analysis indicated a bidirectional causal relationship between the GM and insomnia. We have predicted potential TCMs that act on GM to intervene in insomnia. The findings of this research offer valuable perspectives on the mechanism and clinical investigation of insomnia caused by GM.