Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
ONCOLOGY / RESEARCH PAPER
Development and validation of a prognostic nomogram for lower-grade glioma based on an autophagy-related lncRNA signature
1
The First Affiliated Hospital of Shantou University Medical College, China
2
Cell Biology Department, Wuxi School of Medicine, Jiangnan University, China
Submission date: 2021-10-15
Final revision date: 2021-12-15
Acceptance date: 2021-12-31
Online publication date: 2022-01-01
Corresponding author
Wei-jiang Zhao
Cell Biology Department, Wuxi School of Medicine, Jiangnan University, No.1800 Lihu Dadao, Binhu District, 214122, Wuxi, China
Cell Biology Department, Wuxi School of Medicine, Jiangnan University, No.1800 Lihu Dadao, Binhu District, 214122, Wuxi, China
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Gliomas account for 75% of the primary malignant brain tumors. The prognosis and treatment planning vary in lower-grade gliomas (LGG) due to their heterogeneous clinical behaviors. The dysregulation of autophagy-related (ATG) lncRNAs plays a crucial role in LGG. We aimed to develop and validate an ATG lncRNA risk signature, and a survival nomogram with integration of novel prognostic for LGG patients.
Material and methods:
Differentially expressed ATG lncRNAs were screened out based on TCGA and GTEx RNA-seq databases. ATG lncRNA prognostic signature was then established by Kaplan–Meier, univariate Cox proportional hazards regression, Least absolute shrinkage and selection operator (LASSO) regression and multivariate Cox proportional hazards regression, with its predictive value validated by time-dependent receiver operating characteristic (ROC) curves. Kaplan–Meier, univariate Cox regression and multivariate Cox proportional hazards regression were used to screen out clinical and molecular variables. A nomogram was developed and internally validated by ROC and calibration plots.
Results:
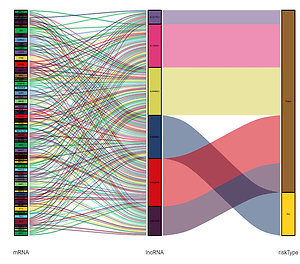
An ATG lncRNA risk signature was constructed with six differentially expressed lncRNAs (LINC00599, LINC02609, AC021739.2, AL118505.1, AL354892.2, and AL590666.2). Based on the risk signature, a nomogram was developed by addition of the significant prognostic clinical variables (age and grade) and molecular variables (IDH status and MGMT status).
Conclusions:
We identified an ATG lncRNA risk signature and develop a nomogram for individualized survival prediction in LGG patients. A user-friendly free online calculator to facilitate the use of this nomogram among clinicians is also provided: https://linstu2009.shinyapps.io/LGGPRODICTORapp/?_ga=2.3154800.1506830296.1588641469-159983587.1588641469.
Gliomas account for 75% of the primary malignant brain tumors. The prognosis and treatment planning vary in lower-grade gliomas (LGG) due to their heterogeneous clinical behaviors. The dysregulation of autophagy-related (ATG) lncRNAs plays a crucial role in LGG. We aimed to develop and validate an ATG lncRNA risk signature, and a survival nomogram with integration of novel prognostic for LGG patients.
Material and methods:
Differentially expressed ATG lncRNAs were screened out based on TCGA and GTEx RNA-seq databases. ATG lncRNA prognostic signature was then established by Kaplan–Meier, univariate Cox proportional hazards regression, Least absolute shrinkage and selection operator (LASSO) regression and multivariate Cox proportional hazards regression, with its predictive value validated by time-dependent receiver operating characteristic (ROC) curves. Kaplan–Meier, univariate Cox regression and multivariate Cox proportional hazards regression were used to screen out clinical and molecular variables. A nomogram was developed and internally validated by ROC and calibration plots.
Results:
An ATG lncRNA risk signature was constructed with six differentially expressed lncRNAs (LINC00599, LINC02609, AC021739.2, AL118505.1, AL354892.2, and AL590666.2). Based on the risk signature, a nomogram was developed by addition of the significant prognostic clinical variables (age and grade) and molecular variables (IDH status and MGMT status).
Conclusions:
We identified an ATG lncRNA risk signature and develop a nomogram for individualized survival prediction in LGG patients. A user-friendly free online calculator to facilitate the use of this nomogram among clinicians is also provided: https://linstu2009.shinyapps.io/LGGPRODICTORapp/?_ga=2.3154800.1506830296.1588641469-159983587.1588641469.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.