Introduction
Acute myeloid leukaemia (AML), the most common acute leukaemia in adults, is associated with short survival and is characterized by specific genomic abnormalities, for instance chromosomal aberrations and DNA mutations with prognostic impact, such as FLT3 (fms-like tyrosine kinase 3), NPM1 (Nucleophosmin 1) CEBPA, and recently DNMT3A (DNA methyltransferase 3a) R882 mutations [1–4].
Telomerase reverse transcriptase (TERT) protein, encoded by the TERT gene, plays an important role in maintaining telomere stability and its length, and therefore it helps in preserving chromosomal stability [5]. Associations between telomere length, disease progression, overall survival (OS), and prognosis have already been reported in haematological cancers [5–7].
Previous studies revealed that the presence of single nucleotide functional polymorphisms (SNPs) in the human TERT gene may affect its expression and telomerase activity, and may contribute to the development of cancer as well as the outcome of chemotherapy [8–10]. We previously reported that TERT rs2736100 predisposes to all BCR-ABL-negative myeloproliferative neoplasms (MPN), regardless of the clinical or molecular subtype [10]. Similarly, some TERT promoter SNPs were associated with an increased risk of developing haematological malignancies like acute lymphoblastic leukaemia [11] and chronic lymphocytic leukemia [12]. A meta-analysis showed that rs2853669 alone did not increase or decrease the overall cancer risk or influence the prognosis [13]. Conflicting results in the meta-analyses [9, 13] may be due to ethnicity and pathology selection, as they contained only one or two studies on AML.
Recently, two TERT gene SNPs, namely rs2736100 and rs2853669, previously associated with myeloproliferative neoplasms [10, 14–16], emerged as possible predisposing factors to AML and as prognostic markers for survival [17, 18]. In contrast, Krahling et al. [19] did not find TERT rs2736100 to represent a susceptibility factor for chronic myeloid leukaemia and AML cases in the Hungarian population.
Whereas these studies are the only ones that examined the association between rs2736100 and rs2853669 in TERT gene and the risk of AML, additional studies are necessary to investigate this possible association in groups of various ethnic origins.
We aimed to investigate the contribution of TERT rs2736100 and rs2853669 SNPs in defining the genetic predisposition to AML in Romanian patients – an Eastern European population – their associations with different prognostic markers, and their impact on survival, outcome, and prognosis of patients. In addition, we analysed the association of TERT SNPs in AML in the presence or absence of DNMT3A, NPM1, and FLT3 mutations.
Material and methods
Patients and controls
Our study comprised 509 participants (240 males, 269 females), consisting of 146 AML patients and 363 healthy participants. The present study included consecutive AML patients from the central region of Romania. Samples were collected at the moment of diagnosis at admission in the Haematology Clinics from Tîrgu Mureş, Romania before starting chemotherapy. The inclusion criteria were as follows: patients diagnosed with AML, adults (> 18 years old), and having given written informed consent. Briefly, the therapy protocols used for AML induction were the “5+2”, “7+3” regimen and all-trans-retinoic acid (ATRA) for LAM M3 (acute promyelocytic leukaemia). The doses were adjusted mainly according to the patient’s age (standard or low dose).
The controls were unrelated healthy individuals, from the same geographic area, without any malignancy, and they were included in control group in parallel with the patients. Controls were referred to the Emergency County Hospital from Tîrgu Mureş for investigation, mainly due to anaemia or leukocytosis. The 2 groups of subjects had a similar gender distribution.
Our study was approved by the Ethical Committee of the University of Medicine and Pharmacy of Tîrgu Mureş, Romania and was performed in accordance with the Declaration of Helsinki.
Methods
Genomic DNA (gDNA) was extracted from fresh blood using DNA Quick-gDNA MiniPrep (ZymoResearch) and PureLink Genomic DNA Mini (Invitrogen) kits.
TERT rs2736100 (c.1574-3777G>T) and rs2853669 (c.-245T>C) polymorphisms were genotyped by TaqMan SNP genotyping assays (c_1844009_10, c_8773290_10) from ThermoFisher Scientific according to the manufacturer’s instructions and run on a 7500 Fast Dx Real-Time PCR Instrument (Applied Biosystems).
FLT3 (ITD, D835 c.2503G>A/C/T, c.2504A>C/T), DNMT3A (R882 c.2644C>A/G/T and C.2645G>A), and NPM1 (c.863_864insTCTG, type A) mutation status was analysed in each AML case as previously reported [20–22]. Randomly, in 10% of AML cases, FLT3 and DNMT3A were also investigated by HRM (in-house method) while NPM1 c.863_864insTCTG mutation was analysed by castPCR.
Statistical analysis
Quantitative continuous variables were expressed as mean (± standard deviation) for Gaussian distribution and as median and interquartile range (percentile 25%; percentile 75%) for data with deviations from normal distribution of probability. Qualitative variables were described by absolute frequencies (number of cases) and relative frequencies (%). The χ2 test was used in bivariate analysis of associations between qualitative variables. The impact of TERT rs2736100 and rs2853669 polymorphisms on AML risk was tested using logistic regression analysis, the size effect of association being quantified by odds-ratio (OR) and 95% confidence level for OR.
The hypothesis of difference in survival time distribution was investigated by non-parametric Kaplan-Meier method while the univariate and multivariate effects of TERT rs2736100 and TERT rs2853669 SNPs on OS time in AML patients were tested using Cox regression analysis. The effect of the studied SNPs on OS was estimated adjusting for the following covariates: age group, gender, cytogenetic risk group, ECOG status, FLT3, DNMT3A or NPM1 mutation, AML subtype (de novo; secondary or therapy-related), and treatment (high dose of chemotherapy, low dose of chemotherapy, or high dose + haematopoietic stem cell transplantation). Because the type of treatment did not accomplish the condition of proportional hazard ratio, we used a stratified Cox regression analysis. The effect size of association between each of the studied SNPs and OS was quantified by hazard ratio (HR) adjusted according to the above-mentioned factors. Candidate predictors included in multivariable Cox regression models were significant univariate predictors along with all important clinical variables [23], so that the number of predictors (p) was lower than m/15, where m was the number of deaths in the studied sample [24]. Statistical analysis was performed using the advanced environment for statistical computing R version 3.4.4.
Results
The AML group included 74 females and 72 males, while the control group consisted of 195 females and 168 males. In the investigated groups a similar frequency was observed regarding females and males (p = 0.535). The mean age ± SD of studied AML patients at diagnosis was 52.65 ±16.7 years (range: 19–84 years old). In the AML group 84 (57.5%) cases were over 50 years of age. The characteristics of the AML cases included in our study, which consist of demographic, clinical, and biological data and AML classification based on WHO recommendations [25] are presented in Table I.
Table I
Characteristics of AML patients
| Variables | Absolute frequencies (relative frequencies %) |
|---|---|
| Age [years]: | |
| < 65 | 103 (70.5) |
| ≥ 65 | 43 (29.5) |
| Gender: | |
| Female | 74 (50.7) |
| Men | 72 (49.3) |
| WBC [× 103/mm3]: | |
| < 50000 | 123 (84.2) |
| ≥ 50000 | 23 (15.8) |
| PLT [× 103/mm3]: | |
| < 40000 | 69 (47.3) |
| ≥ 40000 | 77 (52.7) |
| Hgb [g/dl]: | |
| < 10 | 99 (67.8) |
| ≥ 10 | 47 (32.2) |
| LDH level [IU/l]: | |
| < 600 | 71 (48.6) |
| ≥ 600 | 75 (51.4) |
| Blasts (in bone marrow, %): | |
| < 70 | 86 (58.9) |
| ≥ 70 | 60 (41.1) |
| Cytogenetic risk: | |
| Low risk (favourable prognostic) | 16 (11.0) |
| Intermediate | 74 (50.7) |
| High risk (Adverse) | 44 (30.1) |
| Not available (NA) | 12 (8.2) |
| Normal karyotypea | 62 (42.5) |
| French-American-British (FAB) classification: | |
| M0 – minimally differentiated AML | 15 (10.3%) |
| M1 – AML without maturation | 25 (17.1%) |
| M2 – AML with maturation | 34 (23.3%) |
| M3 – acute promyelocytic leukaemia | 10 (6.8%) |
| M4 – acute myelomonocytic leukaemia | 27 (18.5%) |
| M5 – acute monocytic leukaemia | 23 (15.8%) |
| M6 – erythroleukemia, M7 – megakaryoblastic, unclassified leukaemia | 12 (8.2%) |
| AML classification based on WHO 2016 [18]: | |
| AML with recurrent cytogenetic anomalies | 33 (22.6%) |
| AML dysplasia related | 44 (30.14%) |
| Therapy-related myeloid neoplasms | 4 (2.74%) |
| AML not otherwise specified | 65 (44.52%) |
| ECOG performance status: | |
| ≤ 1 | 21 (14.4) |
| ≥ 2 | 125 (85.6) |
| ECOG performance status: | |
| 0 | 0 |
| 1 | 21 (14.4) |
| 2 | 18 (12.3) |
| 3 | 65 (44.5) |
| 4 | 42 (28.8) |
| FLT3 status (ITD+D835): | |
| Wild-type | 118 (80.8) |
| Mutant | 28 (19.2) |
| DNMT3A R882 status: | |
| Wild-type | 123 (84.2) |
| Mutant | 23 (15.8) |
| NPM1 c.863_864insTCTG status: | |
| Wild-type | 125 (85.6) |
| Mutant | 21 (14.4) |
| Treatment: | |
| High dose | 79 (54.1) |
| Low dose | 51 (34.9) |
| High dose + haematopoietic stem cell transplantation | 16 (11.0) |
| Response: | |
| Complete remission | 15 (10.3) |
| Partial remission | 24 (16.4) |
| Relapse | 35 (24.0) |
| Resistant | 55 (37.7) |
| No response/Induction death | 17 (11.6) |
We included our patients in the following AML subtypes: 112 (76.7%) with de novo AML, 30 (20.5%) with secondary AML (sAML, arising from myelodysplastic syndrome MDS or from MPN), and 4 (2.7%) with therapy-related AML (tAML for patients with prior chemotherapy). FLT3 mutation was present in 28 of our AML cases, as follows: 23 (15.8%) with ITD mutation, while D835 mutation was present in 8 (5.5%) cases, and 5 AML patients presenting both mutations. Among the AML cases, 74 patients reached remission (15 complete remission CR), 55 cases were chemotherapy resistant, while no response or induction death was observed in 17 patients.
In the AML and control groups the TT, TC, and CC TERT rs2853669 genotypes were 59 (40.4%), 65 (44.5%), 22 (15.1%) vs. 166 (45.7%), 162 (44.6%), 35 (9.6%), respectively. For TERT rs2736100 SNP the AA, AC, and CC genotype distributions in AML and healthy controls were as follows: 31 (21.2%), 76 (52.1%), and 39 (26.7%) vs. 95 (26.2%), 191 (52.6%), and 77 (21.2%). None of the TERT rs2853669 and rs2736100 genotypes demonstrated deviation from the Hardy-Weinberg equilibrium (HWE) in both AML cases and controls (TERT rs2736100: Χ2[2] = 1.10, p = 0.295 in controls, and Χ2[2] = 0.29, p = 0.593 in AML group; TERT rs2853669: Χ2[2] = 0.25, p = 0.617 for the control group and Χ2[2] = 0.34, p = 0.558 for the AML group).
In the overall analysis, TERT rs2736100 and rs2853669 polymorphisms were not associated with AML risk in the codominant, dominant, and recessive models in our population (p > 0.05).
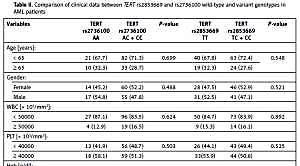
We compared the baseline characteristics of AML patients carrying the variant genotypes for TERT rs2736100 (AC or CC) and for TERT rs2853669 (TC or CC) and patients with wild-type genotypes (Table II). We found no associations between the investigated SNPs and age, gender, white blood cell (WBC) counts, platelets, haemoglobin, lactate dehydrogenase (LDH) levels, percentage of blasts in bone marrow, and cytogenetic risk groups in our cases at diagnosis. Patients with polymorphic TERT rs2736100 (AC or CC) frequently had haemoglobin (Hgb) levels below 10 g/dl, but the difference was not statistically significant (p = 0.084). No association was observed between the variant genotypes and response to treatment (complete, partial remission, relapse, chemotherapy resistant, or induction death) or AML subtype. Likewise, we noticed no association between the studied SNPs and Eastern Cooperative Oncologic Group Scale (ECOG) status (p > 0.05).
Table II
Comparison of clinical data between TERT rs2853669 and rs2736100 wild-type and variant genotypes in AML patients
As a next step, we analysed the distribution of the two TERT rs2853669 and rs2736100 polymorphisms taking into account the somatic mutations displayed in NPM1, FLT3, and DNMT3A genes in AML patients. Interestingly, we noticed no association between NPM1, FLT3, and DNMT3A mutation status and variant genotypes of the investigated SNPs. We looked for the effect of the variant genotype of the investigated SNPs in the presence of at least 1 mutation (DNMT3A, FLT3, NPM1), 2 mutations (FLT3 + NPM1, FLT3 + DNMT3A, DNMT3A + NPM1), or all 3 somatic mutations in AML cases, but no association was observed (p > 0.05 for all comparison performed; data not shown).
The results of Kaplan-Meier analysis (Table III) showed that the distribution of survival time was different for the following factors: age, cytogenetic risk, treatment, and FLT3 mutation, respectively. We found no significant differences in survival time regarding TERT rs2736100 or rs2853669 genotypes in AML, nor for NPM1 type A, DNMT3A R882 mutation, and AML subtypes (de novo, sAML, tAML). Furthermore, we investigated the survival time according to the ECOG status and we noticed a shorter survival in patients with ECOG 4 status, the OS median being 5 months (95% CI: 0.8–9.2).
Table III
Univariate analysis of overall survival time in patients with AML: Kaplan-Meier method
| Factors | Median of overall survival time (95% CI) | P-value* |
|---|---|---|
| Age category: | ||
| < 65 years | 9.0 (7.3–10.7) | |
| ≥ 65 years | 3.0 (1.6–4.4) | < 0.001 |
| < 50 years | 10.0 (8.0–11.9) | |
| ≥ 50 years | 5.0 (3.1–6.9) | 0.001 |
| Cytogenetic risk: | ||
| Low risk (favourable prognostic) | 10.0 (4.7–15.3) | |
| Intermediate | 8.0 (6.2–9.8) | |
| High risk (adverse) | 3.0 (1.5–4.5) | 0.001 |
| Treatment: | ||
| High dose | 8.0 (6.2–9.8) | |
| Low dose | 4.0 (2.5–5.4) | |
| High dose + haematopoietic stem cell transplantation | 12.0 (9.1–14.9) | < 0.001 |
| FLT3 mutation (ITD + D835): | ||
| Negative | 8.0 (6.7–9.3) | |
| Positive | 2.0 (0.0–4.3) | 0.001 |
| NPM1 type A mutation: | ||
| Negative | 7.0 (5.7–8.3) | |
| Positive | 5.0 (1.9–8.0) | 0.310 |
| DNMT3A mutation: | ||
| Negative | 8.0 (6.8–9.2) | |
| Positive | 4.0 (2.3–5.7) | 0.290 |
| TERT rs2736100: | ||
| AA | 8.0 (4.8–11.2) | |
| AC | 5.0 (1.9–8.0) | |
| CC | 8.0 (5.5–8.5) | 0.108 |
| TERT rs2853669: | ||
| TT | 8.0 (6.4–9.6) | |
| CT | 4.0 (1.6–6.4) | |
| CC | 9.0 (5.8–12.2) | 0.315 |
The results of multivariate Cox regression showed that TERT rs2853669 SNP was a significant predictor for overall survival in AML patients (Table IV). After adjusting for the main prognostic factors in AML, such as age, gender, cytogenetic risk group, FLT3, DNM3A or NPM1 mutations, AML subtype, and treatment, the estimated adjusted hazard ratio (HR adjusted = 1.54, 95% CI: 1.01–2.35) showed that the TERT rs2853669 variant genotype (CC + CT) had a negative influence on survival time. We evaluated the predictive accuracy of Cox-regression-tested models. Based on the concordance index (C-index), all regression-tested models demonstrated a good discriminant accuracy, with an estimated value of the concordance index between 0.70 and 0.80.
Table IV
Associations between TERT SNPs and overall survival of AML patients
| Gene polymorphisms | Patients | Death | MST | HRa (95% CI) | P-valueb | Model performance (C-statistics; likelihood ratio test) |
|---|---|---|---|---|---|---|
| TERT rs2736100: | ||||||
| AA | 31 (21.23) | 26 | 8.00 | 1.00 (Reference) | – | C = 0.73;Χ;Χ2(10) = 29.79; p = 0.0009 |
| AC | 76 (52.05) | 69 | 5.00 | 1.40 (0.72–0.83) | 0.206 | |
| CC | 39 (26.71) | 31 | 8.00 | 0.99 (0.54–1.84) | 0.985 | |
| AC + CC | 115 (78.77) | 100 | 7.00 | 1.27 (0.76–2.10) | 0.358 | C = 0.72; Χ2(9) = 27.48; p = 0.0012 |
| AA + AC | 107 (73.29) | 95 | 6.00 | 1.00 (Reference) | – | C = 0.72; Χ2(9) = 27.71; p = 0.0011 |
| CC | 39 (26.71) | 31 | 8.00 | 0.77 (0.48–1,22) | 0.264 | |
| TERT rs2853669: | ||||||
| TT | 59 (40.41) | 49 | 8.00 | 1.00 (Reference) | – | C = 0.73;Χ;Χ2(10) = 34.32; p = 0.0002 |
| CT | 65 (44.52) | 61 | 4.00 | 1.74 (1.12–2.70) | 0.014+ | |
| CC | 22 (15.07) | 16 | 9.00 | 1.03 (0.52–2.03) | 0.932 | |
| CT + CC | 87 (59.59) | 77 | 7.00 | 1.54 (1.01–2.35) | 0.043+ | C = 0.72; Χ2(9) = 31.43; p = 0.0002 |
| CT + TT | 22 (15.07) | 16 | 9.00 | 1.00 (Reference) | – | C = 0.71; Χ2(9) = 26.98; p = 0.0014 |
| CC | 22 (16.42) | 16 | 8.00 | 0.77 (0.41–1.56) | 0.427 | |
Discussion
In our study, we analysed the association of rs2853669 and rs2736100 in the TERT gene with the risk of developing AML, in relation to clinical and prognostic markers and to OS in AML. Although there was no significant difference between TERT rs2853669 and rs2736100 SNPs variant alleles or genotypes in AML cases, an increasing TERT rs2853669 homozygous variant frequency trend was noted (p = 0.067).
Our results are different from those reported earlier in South-Eastern Swedish AML patients [17], where the variant CC genotype of TERT rs2853669 was strongly associated with an increased risk for AML (p = 0.00015) and with a decreased OS [17]. In the same study, TERT rs2736100 showed an association with the risk for developing AML (p = 0.03), but without any effect on survival in AML. Regarding the relation between rs2736100 and AML overall survival, we observed partially overlapping results with those reported by Mosrati et al. [17].
The study performed by Krahling et al. [19] on 308 cases with AML (195 de novo cases, 93 myelodysplasia-related AML cases, and 20 therapy-related AML cases) revealed no association between TERT rs2736100 SNP and AML susceptibility, similarly to what we noticed but in contrast to the results of Mosrati et al. [17]. A possible explanation for these discordant results may be the origin of the studied groups: Mosrati et al. [17] investigated cases from Northern Europe while Krahling et al. [19] analysed individuals from Eastern Europe, as our study did.
Our association analysis revealed no significant correlations between TERT rs2853669 and rs2736100 polymorphisms and the risk of AML in our Romanian population, so our findings do not support the hypothesis of their involvement in the aetiology of AML in the Romanian population.
Furthermore, by applying univariate analysis (Kaplan-Meyer method) we found no association between TERT rs2853669 or TERT rs2736100 variant genotypes and AML subtype, cytogenetic risk group, OS, as well as response to treatment. We found no significant differences regarding survival time (p = 0.504) or AML predisposition (p = 0.535) between genders in the AML group.
We investigated the combined effect of both TERT rs2853669 and rs2736100 polymorphisms (the presence of both variant genotypes) and noticed no association with the risk of developing AML, and we considered that this association is not a predictor for AML risk (p = 0.671). In addition, our findings revealed that the combined variant genotypes of the investigated SNPs did not modify overall survival in AML patients (p = 0.223).
We have investigated the OS time in AML and whether TERT rs2853669 and rs2736100 were independent prognostic markers for OS in their AML cohort by performing multivariate analysis using Cox regression tested models. Multivariate analysis showed that the variant rs2853669 CT and CT + CC genotypes were predictors for patient survival in AML cases (p = 0.014; p = 0.043), after adjusting for the main prognostic factors in AML, such as age, gender, cytogenetic risk group, FLT3, DNM3A, or NPM1 mutations. The effect size of association between rs2853669 and OS quantified by hazard ratio was 1.74, 95% CI: 1.12–2.70 for CT genotype and 1.54, 95% CI: 1.01–2.35 for CT + CC (dominant model). Our observations are in line with those reported by Mosrati et al. [17], which underlined that the TERT rs2853669 CC genotype may represent an independent biomarker associated with poor prognosis in AML. Multivariate Cox-regression analysis performed in our study indicated that TERT rs2736100 did not represent a risk factor for AML development. These results are consistent with previously reported data [17]. Multivariate analysis was not performed by Krahling et al. [19] in their investigation of OS time and whether TERT rs2853669 and rs2736100 were prognostic markers for OS in their AML cohort.
Taking into account that the Concordance index (C-index or C-statistic) obtained was between 0.70 and 0.80, we consider that we had strong models and that the regression test models used demonstrated good discriminant accuracy.
There are several limitations of our study that should be mentioned: telomerase activity and telomere length were not evaluated in AML cases, the lack of investigation of CEBPA and RUNX1 gene mutations, the relatively small study sample size, and the lack of gene-environment interaction evaluation.
This is the first study focusing on the impact of interaction between TERT gene polymorphisms and DNMT3A mutations on AML risk. In our AML cohort, TERT rs2853669 and rs2736100 variant genotypes were not associated with DNMT3A mutations. We observed that DNMT3A R882 affected the patients’ OS (OS median was 4 months in cases with DNMT3A mutation).
Considering the low number of studies that investigated the association between TERT polymorphisms and the risk of acute myeloid leukaemia, and the fact that the findings are still preliminary and inconclusive, further research is necessary in other ethnic groups.
In conclusion, our results indicated that TERT rs2853669 and rs2736100 polymorphisms were not risk factors for developing AML in the Romanian population, but the presence of TERT rs2853669 CT or CT + CC variant genotypes had a negative effect on AML patients’ overall survival in the presence of other known prognostic factors.