Introduction
Breast cancer is the most frequently diagnosed cancer in women, accounting for 11.7% of all newly diagnosed cancer cases in 2020 [1]. Timely detection and treatment are necessary to improve the treatment response and prognosis of patients with breast cancer. However, breast cancer has a higher mortality rate owing to late diagnosis, chemotherapy resistance, and metastasis [2, 3]. Breast cancer is a highly heterogeneous disease with unique and complex histopathological patterns and clinical behaviors [4, 5]. Moreover, breast cancer pathogenesis is complex and poorly understood. Several studies have reported specific tumor markers for the evaluation of cancer progression [6]. Analysis of biomarkers is essential for the early diagnosis and adequate treatment of breast cancer.
Circular RNAs (circRNAs) have gained increasing attention owing to their involvement in cancer progression. CircRNAs are special non-coding RNAs with closed-loop structures that lack 5′- and 3′-ends [7]; however, their specific roles in diseases remain ambiguous. Deregulation of circRNAs is associated with the development and progression of various diseases, including cancer [8, 9]. Previous studies have reported several circRNAs with potential diagnostic value, whose aberrant expression is associated with advanced tumor stages and poor prognosis and therapeutic response in patients with breast cancer [10–12]. Moreover, several circRNAs have been reported to modulate cancer cell phenotypes and functions, such as proliferation, apoptosis, energy metabolism, and metastasis [13, 14]. Several circRNAs have been identified owing to the advancement in RNA sequencing technology; however, the functions of most circRNAs remain unknown. Hsa_circ_0119412 and circRNA_102958 levels were initially reported to be upregulated in gastric cancer using circRNA microarray data [15]. We also observed increased circRNA_0119412 expression levels in breast tumor tissues in our study, suggesting that circ_0119412 may be involved in breast cancer development. However, the specific functions and underlying molecular mechanisms of circ_0119412 in breast cancer remain unclear.
CircRNAs containing microRNA (miRNA) response elements may function as sponges of their target miRNAs to modulate their expression [16]. Advancements in bioinformatics have facilitated the prediction of miRNAs bound to circRNAs [17]. In this study, we determined the functional mechanisms of circ_0119412 by assessing its target miRNA, miR-1205. miR-1205 expression is downregulated in various cancers [18, 19]. miRNAs modulate the expression of genes by targeting their 3′-untranslated regions (UTRs) [20]. Bioinformatics analysis has revealed that miR-1205 contains complementary sequences of polypeptide N-acetylgalactosaminyltransferase 6 (GALNT6) 3′-UTR, suggesting that miR-1205 may mediate GALNT6 silencing. GALNT6 mediates the initial steps of mucin-type O-glycosylation [21]. GALNT6 also promotes carcinogenesis in various cancers [21–23]. However, the mechanism by which the interaction of GALNT6 with miR-1205 affects the development of breast cancer remains unknown.
In this study, we determined the expression levels and functions of circ_0119412 in breast cancer cells and tumor specimens of an animal model. We also assessed the associations of miR-1205 with circ_0119412 and GALNT6 and their influence on breast cancer development. Our study aimed to determine the anti-proliferative effect of circ_0119412 knockdown on breast cancer cells. To the best of our knowledge, this study is the first to examine the roles of circ_0119412 in breast cancer.
Material and methods
Tissue specimens
Tumor (n = 35) and normal (non-cancerous; n = 35) specimens were collected during surgery from the female patients with breast cancer enrolled in our hospital. All patients provided written consent prior to their participation in this study. Inclusion criteria were as follows: 1) patients diagnosed with breast cancer and 2) patients or their family members providing written consent. Exclusion criteria were as follows: 1) patients without a history of breast cancer, 2) patients who underwent therapy before surgery, and 3) patients with other diseases. This study was designed and conducted with approval from our hospital. Characteristics of all patients with breast cancer included in this study are listed in Table I.
Cell lines and cell culture
Common breast cancer (MDA-MB-231, MCF-7, and MDA-MB-468) and normal breast epithelial (MCF-10A) cell lines were purchased from Procell (Wuhan, China). MCF-7 and MCF-10A cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, USA) containing 10% fetal bovine serum (FBS). MDA-MB-468 and MDA-MB-231 cells were cultured in Leibovitz’s L-15 medium (Gibco) containing 10% FBS. Cultures were incubated at 37°C and 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using the Trizol reagent and analyzed using NanoDrop2000 (Thermo Fisher Scientific, USA). Then, total RNA was reverse-transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, USA) and miRNA Reverse Transcription Kit (Qiagen, Germany), according to the manufacturers’ protocols. cDNA was then amplified via qPCR using the QuantiTect SYBR Green PCR Kit (Qiagen) and LightCycler 96 thermocycler (Roche, Switzerland). Relative expression levels of genes were normalized against uracil 6 (U6) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels using the 2-ΔΔCT method. All primer sequences used in this study are listed in Table II.
Table II
Real-time PCR primer sequences
Subcellular localization
The Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, Canada) was used to isolate RNAs from the cytoplasm and nucleus, and enrichment of circ_0119412 in each fraction was assessed using RT-qPCR. As GAPDH is mainly located in the cytoplasm and U6 is mainly located in the nucleus, GAPDH was used as the cytoplasmic control, and U6 was used as the nuclear control.
RNase R treatment
RNA samples were treated with RNase R (2 U/µg; BioVision, USA) and subjected to RT-qPCR to assess the changes in the expression levels of circ_0119412 and the linear gene period circadian regulator 2 (PER2).
Cell transfection
Oligonucleotides for circ_0119412 silencing (si-circ) and its matched negative control (si-NC) were obtained from GenePharma (China). miR-1205 mimic, miR-1205 inhibitor, mimic-NC, and inhibitor-NC were purchased directly from RiboBio (China). Oligonucleotides against GALNT6 (si-GALNT6) and its matched negative control (si-NC) were also purchased from GenePharma. Then, the oligonucleotides were transfected individually or in specific combinations into the experimental cells using the Lipofectamine 3000 reagent (Invitrogen, China). After 24 h, cells were collected and the transfection efficiency was determined via RT-qPCR and western blotting.
Cell counting kit (CCK)-8 assay
Transfected cells were seeded in a 96-well plate at a density of 5,000 cells/well. After culturing at 37°C for 0, 24, 48, and 72 h, the cells in each well were mixed with the CCK-8 reagent (Invitrogen) and maintained for another 4 h. Cell viability was determined based on the absorbance at 450 nm using a microplate reader (Bio-Rad).
Transwell assay
Transwell chambers (Corning, USA) pretreated with Matrigel (Corning) were used for cell invasion analysis. Transfected cells were cultured for 24 h, collected in a serum-depleted medium, and transferred to the top transwell chambers. A fresh culture medium containing 20% FBS was added to the bottom chamber. Cells were allowed to invade the membrane for 24 h. Cells that invaded the lower surface of the membranes were immobilized with methanol and stained with crystal violet. Cell morphology was observed under a light microscope (Leica, Germany).
Flow cytometry assay
Transfected cells were cultured for 48 h and collected after trypsin digestion. An Annexin V-FITC/PI Apoptosis Detection Kit (Vazyme, China) was used to identify the apoptotic cells. Briefly, cells were resuspended in Annexin V-FITC binding buffer and stained with annexin V-FITC and propidium iodide. Then, a flow cytometer (Beckman Coulter, USA) was used to estimate the cell apoptosis rate.
Animal study
Six-week-old female nude mice were purchased from Beijing Charles River Laboratories (China). Geneseed (China) assembled the lentiviruses of short hairpin RNAs targeting circ_0119412 (sh-circ) or GALNT6 (sh-GALNT6) and miR-1205 overexpression (Lv-miR-1205) and their negative controls (sh-NC and Lv-NC, respectively). MCF-7 cells were transfected with either sh-circ or sh-NC lentiviral vectors, and 2 × 106 cells were subcutaneously injected into each nude mouse (n = 5 per group) to induce tumor formation. Tumor volume (length × width2 × 0.5) was recorded weekly for 5 weeks, after which the mice were sacrificed to excise the tumors for analyses. All animal experimental procedures used in this study were approved by our hospital.
Dual-luciferase reporter assay
Mutant (MUT) and wild-type (WT) circ_0119412 reporter plasmids were generated using the pmirGLO plasmid (Promega, USA). Moreover, WT and MUT GALNT6 reporter plasmids were constructed in a similar manner. As miR-1205 and GALNT6 share two binding sites, various GALNT6 reporter plasmids, namely WT, Co-MUT (mutations at two sites), MUT1 (mutation at one site), and MUT2 (mutation at the other site) plasmids, were constructed and transfected with the miR-1205 or NC mimic into MDA-MB-231 and MCF-7 cells. After 48 h, luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega).
RNA immunoprecipitation (RIP) assay
The Imprint RNA Immunoprecipitation Kit and experimental (anti-IgG and anti-Ago2) antibodies were acquired from Sigma-Aldrich (USA). MDA-MB-231 and MCF-7 cells were treated with the RIP lysis buffer, and cell lysates were incubated with Protein A Magnetic Beads pre-coated with anti-IgG or anti-Ago2. Then, RNA complexes were washed and purified. Finally, RT-qPCR was performed to quantify the expression levels of miR-1205 and circ_0119412.
Western blotting
Total protein was extracted from the cells using the radioimmunoprecipitation assay lysis reagent (Beyotime, Shanghai, China). Proteins were quantified using a BCA kit (Beyotime). After separation using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the protein bands were transferred to polyvinylidene fluoride membranes (Beyotime), blocked with 5% skim milk for 1 h at room temperature, and incubated with primary antibodies against GAPDH (1 : 2500 dilution; ab181602; Abcam, USA) and GALNT6 (1 : 1000 dilution; ab151329; Abcam) at 4°C overnight, followed by incubation with secondary antibodies (1 : 10000 dilution; ab205718; Abcam) for 2 h at room temperature. Finally, an ECL kit (Beyotime) was used to visualize the protein bands.
Statistical analysis
Each assay was performed in triplicate. GraphPad Prism v7.0 (GraphPad Software, USA) was used to process the data and construct the graphs. Pearson’s rank correlation was used to ascertain the correlation between groups. Student’s t-test was used to compare two groups. Analysis of variance with post-hoc Tukey’s correction was used for multigroup comparisons. Data are presented as the mean ± standard deviation. Statistical significance was set at p < 0.05.
Results
Circ_0119412 expression levels are increased in breast cancer cell lines and tumor specimens
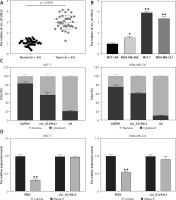
First, we determined circ_0119412 expression in breast cancer. As depicted in Figures 1 A and B, circ_0119412 levels were markedly higher in the breast cancer specimens and (MDA-MB-231, MCF-7, and MDA-MB-468) cell lines than in the normal specimens and non-cancerous (MCF-10A) cell lines (tumor:normal = 2.91, MDA-MB-231:MCF-10A = 3.39, MCF-7:MCF-10A = 3.95, and MDA-MB-468:MCF-10A = 1.57). As MDA-As MB-231 and MCF-7 cells exhibited the highest circ_0119412 expression, they were selected for subsequent experiments. CircRNA subcellular localization analysis revealed that relative to the nucleus, circ_0119412 was mainly and abundantly distributed in the cytoplasm (approximately 60%) of breast cancer cells (Figure 1 C). RNase R was used to digest the total RNA extracted from cells to confirm the presence of circ_0119412. RNase R barely affected the expression of circ_0119412 but substantially decreased the expression of its linear parental gene, PER2 (Figure 1 D). These results indicate that circ_0119412 dysregulation may be associated with breast cancer development.
Figure 1
Increased circ_0119412 expression was detected in the breast cancer cell lines and tumor specimens. A – Circ_0119412 expression in tumoral and normal samples was checked via RT-qPCR. B – The circ_0119412 expression levels in MDA-MB-231, MCF-10A, MCF-7, and MDA-MB-468 cells were quantified by means of RT-qPCR, *P < 0.05, **p < 0.001 vs. MCF-10A. C – The circ_0119412 expression levels within the cytoplasm and nuclei of the cells were determined via RT-qPCR. D – The presence of circ_0119412 was confirmed by treating the RNA with RNase R, **p < 0.001 vs. Control
Circ_0119412 deficiency inhibits aggressive breast cancer development in vitro and in vivo
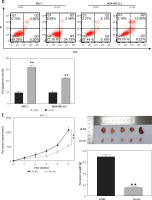
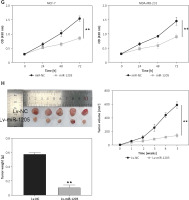
To determine the functions of circ_0119412, we downregulated its expression in MCF-7 and MDA-MB-231 cells. circ_0119412 expression levels were markedly lower in cell lines transfected with si-circ than in those transfected with si-NC (p < 0.001; Figure 2 A). CCK-8 assay revealed lower proliferation rates in MCF-7 and MDA-MB-231 cells transfected with si-circ than in those transfected with si-NC (p < 0.001; Figure 2 B). Moreover, transwell assay revealed considerably lower invasion rates in cancer cells transfected with si-circ than in those transfected with si-NC (p < 0.001, Figure 2 C). However, circ_0119412 silencing reversed these effects (p < 0.001; Figure 2 D). Furthermore, animal models were established by injecting sh-circ- or sh-NC-transfected MCF-7 cells into nude mice. Compared with the sh-NC-administered group, tumor sizes and weights were lower in the sh-circ-administered group (p < 0.001; Figure 2 E). Collectively, these results suggest that circ_0119412 deficiency inhibits breast cancer progression.
Figure 2
Circ_0119412 deficiency blocked breast cancer progression in vitro and in vivo. A – The efficiency of the circ_0119412 silencing was ascertained through RT-qPCR, **p < 0.001 vs. si-NC. B – Circ_0119412 silencing’s effect on the proliferative abilities of the cells was assessed by means of the CCK-8 experiment, **p < 0.001 vs. si-NC. C – The effect of circ_0119412 silencing on cell invasion was assessed by executing the transwell experiment, **p < 0.001 vs. si-NC. D – The effect of circ_0119412 silencing on apoptosis was determined via flow cytometry, **p < 0.001 vs. si-NC. E – The animal study was conducted to assess how circ_0119412 knockdown affected the growth of solid tumors in vivo, **p < 0.001 vs. sh-NC
miR-1205 is a downstream target of circ_0119412
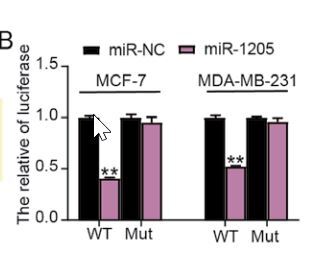
CircInteractome prediction revealed that circ_0119412 possessed an miR-1205-binding site (Figure 3 A). Dual-luciferase reporter assay demonstrated that restoring miR-1205 expression considerably decreased the luciferase activity in MCF-7 and MDA-MB-231 cells containing the circ_0119412 WT reporter plasmid compared to that in cells containing the MUT plasmid (p < 0.001; Figure 3 B). RIP assay revealed that miR-1205 and circ_0119412 were more abundant in the anti-Ago2-enriched RIP group than in the anti-IgG-enriched RIP group (p < 0.001; Figure 3 C). Further expression analysis revealed considerably lower miR-1205 expression levels in breast cancer tumor specimens and cell lines (MDA-MB-231 and MCF-7) than in normal specimens and cell lines (MCF-10A) (tumor : normal = 0.54, MDA-MB-231:MCF-10A = 0.30, and MCF-7 : MCF-10A = 0.46; Figures 3 D and E). Furthermore, miR-1205 expression was inversely correlated with circ_0119412 expression in breast cancer tissues (Figure 3 F). CCK8 assay confirmed that miR-1205 overexpression significantly inhibited cancer cell proliferation in vitro (p < 0.001; Figure 3 G). The in vivo experiment revealed that miR-1205 overexpression reduced the tumor volumes and weights in mice (p < 0.001; Figure 3 H). Overall, these results suggest that circ_0119412 targets miR-1205.
Figure 3
MiR-1205 was a circ_0119412 target. A – CircInteractome predicted that miR-1205 was a potential circ_0119412 target. B, C – MiR-1205’s binding with circ_0119412 was validated via the dual-luciferase reporter assay (B, **p < 0.001 when compared to miR-NC) and RIP assay (C, **p < 0.001 vs. anti-IgG). D – The miR-1205 expression levels in tumoral and normal tissues were measured via RT-qPCR. E – The miR-1205 expression levels in MDA-MB-231, MCF-7, and MCF-10A cells were assessed via RT-qPCR, **p < 0.001 vs. MCF-10A. F – The negative correlation of miR-1205 expression with circ_0119412 expression in tumor tissues. G – miR-1205 overexpression’s effect on the proliferative abilities of the cells was assessed by means of the CCK-8 experiment, **p < 0.001 vs. miR-NC. H – The animal study was conducted to assess how miR-1205 overexpression affected the growth of solid tumors in vivo, **p < 0.001 vs. Lv-NC
Circ_0119412 knockdown inhibits breast cancer development by increasing miR-1205 expression
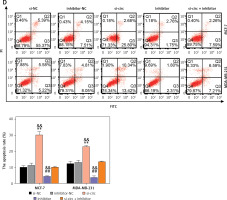
To assess the impact of the interplay between miR-1205 and circ_0119412 on breast cancer progression, rescue experiments were performed using MCF-7 and MDA-MB-231 cells. si-circ, miR-1205 inhibitor, or their combination (si-circ + miR-1205 inhibitor) was transfected into the cells. miR-1205 expression was remarkably elevated by si-circ but decreased by the miR-1205 inhibitor (p < 0.001; Figure 4 A). Moreover, transfection with si-circ + the miR-1205 inhibitor reduced the expression of miR-1205 compared with transfection with si-circ alone. CCK-8 and transwell assays revealed that the proliferative and invasive capacities of MCF-7 and MDA-MB-231 cells were decreased by si-circ and promoted by the miR-1205 inhibitor (p < 0.001, Figures 4 B and C). Furthermore, suppression of cell invasion and proliferation by circ_0119412 knockdown was partially offset by miR-1205 inhibition. In addition, cell apoptosis was promoted by si-circ transfection but suppressed by miR-1205 inhibitor transfection (p < 0.001; Figure 4 D). Increased apoptosis induced by circ_0119412 knockdown was partially repressed by the inhibition of miR-1205. Overall, these results indicate that circ_0119412 knockdown abrogates the malignant behaviors of breast cancer cells via miR-1205 enrichment.
Figure 4
Circ_0119412 knockdown repressed the malignant behavior of breast cancer cells by enriching miR-1205. The rescue experiments were performed using MCF-7 and MDA-MB-231 cells transfected with si-circ, si-NC, miR-1205 inhibitor, inhibitor-NC, or si-circ + miR-1205 inhibitor. A – The miR-1205 expression levels in the transfected cells were assessed via RT-qPCR. B – The proliferative ability of the transfected cells was assessed in the CCK-8 experiment. C – The invasion ability of the transfected cells was studied in the transwell experiment. D – Flow cytometry was carried out to study the apoptosis of the transfected cells. **p < 0.001 vs. si-NC; ##p < 0.001 vs. inhibitor-NC; &&p < 0.01 vs. si-circ + inhibitor
GALNT6 is a target gene of miR-1205
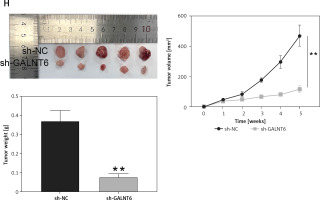
miRDB and TargetScan were used to identify the downstream targets of miR-1205. Gene Expression Profiling Interactive Analysis was used to screen the upregulated genes, with logFC > 2 and adj.p < 0.01. Four genes (cell division cycle-associated 5 [CDCA5], GALNT6, H2B clustered histone 12 [H2BC12, also known as HIST1H2BK], and GATA-binding protein 3 [GATA3]) were found to be overlapping using Venny 2.1.0 (Figure 5 A). Next, we determined their expression levels in clinical tumor specimens. GALNT6 exhibited the highest expression levels in breast cancer tumor specimens (CDCA5: upregulated by 1.13-fold, GALNT6: upregulated by 6.10-fold, HIST1H2BK: upregulated by 1.15-fold, and GATA3: upregulated by 1.23-fold; Figure 5 B); hence, it was chosen for further analyses. TargetScan revealed two miR-1205-binding sites on the GALNT6 3′-UTR sequence fragment (Figure 5 C). GALNT6 WT and MUT reporter plasmids were synthesized and used in a dual-luciferase reporter assay. Combined transfection with the GALNT6 WT reporter plasmid and miR-1205 mimic remarkably diminished the luciferase activity, whereas transfection with the MUT1 + miR-1205 mimic and MUT2 + miR-1205 mimic only partially diminished the luciferase activity in cells (p < 0.001; Figure 5 D). Moreover, co-transfection with the Co-MUT + miR-1205 mimic barely reduced the luciferase activity in cells (p > 0.05, Figure 5 D). GALNT6 mRNA expression levels were markedly higher in MCF-7 and MDA-MB-231 cells than in MCF-10A cells (p < 0.001; Figure 5 E). Notably, miR-1205 expression was negatively associated with GALNT6 mRNA expression in tumor tissues (Figure 5 F). CCK8 assay revealed that GALNT6 knockdown significantly inhibited cell proliferation in vitro (p < 0.001; Figure 5 G). Moreover, GALNT6 knockdown decreased the tumor volumes and weights in vivo (Figure 5 H). These results indicate that GALNT6 is a target gene of miR-1205.
Figure 5
GALNT6 was a miR-1205 target. A – Four genes (CDCA5, GALNT6, HIST1H2BK, and GATA3) were overlapping in miRDB, TargetScan and GEPIA. B – The expression levels of these four genes in tumoral and normal tissues were determined through RT-qPCR C – The binding sites between GALNT6 and miR-1205 were identified using TargetScan. D – The interaction of GALNT6 with miR-1205 was validated via the dual-luciferase reporter experiment, *p < 0.05, **p < 0.001 when compared to WT + NC. E – The GALNT6 expression levels in MDA-MB-231, MCF-7, and MCF-10A cells were quantified via RT-qPCR, **p < 0.001 vs. MCF-10A. F – MiR-1205’s expression in tumors had an inverse correlation with that of GALNT6. G – GALNT6 silencing’s effect on the proliferative abilities of the cells was assessed by means of the CCK-8 experiment, **p < 0.001 vs. si-NC. H – The animal study was conducted to assess how GALNT6 knockdown affected the growth of solid tumors in vivo, **p < 0.001 vs. sh-NC
miR-1205 inhibition aggravates the malignant behaviors of breast cancer cells by increasing GALNT6 expression
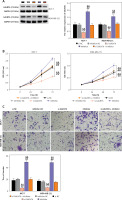
Next, to assess the impact of the interaction between miR-1205 and GALNT6 on breast cancer progression, rescue experiments were performed using MCF-7 and MDA-MB-231 cells. si-GALNT6, miR-1205 inhibitor, or their combination (si-GALNT6 + miR-1205 inhibitor) was introduced into the cells. GALNT6 protein expression levels in MCF-7 and MDA-MB-231 cells were decreased by si-GALNT6 transfection but increased by miR-1205 inhibitor transfection (p < 0.001, Figure 6 A). Moreover, combined transfection with si-GALNT6 + the miR-1205 inhibitor partially enhanced GALNT6 expression compared to the transfection with si-GALNT6 alone. CCK-8 and transwell assays revealed that the proliferative and invasive capacities of cells were considerably decreased in cells transfected with si-GALNT6 alone and increased in cells transfected with the miR-1205 inhibitor (p < 0.001, Figures 6 B and C). Furthermore, repression of cell invasion and proliferation by GALNT6 knockdown was substantially reversed by the inhibition of miR-1205. Cells transfected with si-GALNT6 exhibited enhanced apoptosis, whereas those transfected with the miR-1205 inhibitor exhibited decreased apoptosis (p < 0.001; Figure 6 D). Notably, the effects of GALNT6 knockdown were reversed by miR-1205 inhibition. These results indicate that miR-1205 inhibition aggravates the malignant behaviors of breast cancer cells via the enrichment of GALNT6.
Figure 6
MiR-1205 regulated the cell functions of MCF-7 and MDA-MB-231 by targeting GALNT6. The rescue experiments were executed using MDAMB-231 and MCF-7. Si-GALNT6, si-NC, miR-1205 inhibitor, inhibitor-NC, or si-GALNT6 + miR-1205 inhibitor was transfected into the cells. A – Western blotting was done to estimate the expression of the GALNT6 protein in the transfected cells. B – The proliferation of the transfected cells was evaluated via the CCK-8 experiment. C – The invasion ability of the transfected cells was assessed by executing the transwell experiments. D – Flow cytometry was carried out to evaluate apoptosis among the transfected cells. **P < 0.001 vs. si-NC; ##p < 0.001 vs. inhibitor-NC; &&p < 0.01
Discussion
The main finding of this study was the aberrant overexpression of circ_0119412 in breast cancer cell lines and tumor specimens. We observed that the downregulation of circ_0119412 expression inhibited the malignant phenotypes of cancer cells and decreased the tumor growth in vivo. circ_0119412 was found to regulate breast cancer development by targeting the miR-1205/GALNT6 pathway. This study illustrates the functions of circ_0119412 in breast cancer and suggests its underlying action mechanisms.
Several circRNAs, such as circ_0061825, circ_0025202, and circ_0007294, have been demonstrated to play important roles and affect the growth, invasion, migration, and chemoresistance of breast cancer cells [24–26]. These findings reveal the vital roles of circRNAs in breast cancer tumorigenesis.A previous study reported the increased expression of Circ_0119412 in gastric cancer, and further clinical data suggested its diagnostic value for gastric cancer [15]. Circ_0119412 overexpression has also been reported in colorectal and ovarian cancers, where its downregulation markedly hampered the migration, proliferation, and invasion of cancer cells [27, 28]. To the best of our knowledge, this is the first study to investigate the specific roles of circ_0119412 in breast cancer. Consistent with previous reports, we observed increased expression levels of circ_0119412 in breast cancer tumor specimens in this study. Functional assays revealed that circ_0119412 knockdown repressed the proliferation and invasion, stimulated the apoptosis, and impeded the tumor growth in vivo. Many studies have reported the carcinogenic roles of circ_0119412 in various cancers, suggesting its targeted inhibition as a potential strategy for cancer treatment.
Here, we identified miR-1205 as a downstream target of circ_0119412. miR-1205 expression was negatively correlated with circ_0119412 expression in tumor specimens. Moreover, miR-1205 expression levels were markedly increased in circ_0119412 knockdown breast cancer cells, suggesting that circ_0119412 acts as a sponge and sequesters miR-1205. miR-1205 is also a target of circRNA Ras association domain family member 2 (RASSF2) [29]. miR-1205 enrichment has been reported to alleviate the malignant behaviors of breast cancer cells that are aggravated by circRNA-RASSF2 overexpression [29]. Consistently, we observed decreased expression levels of miR-1205 in breast cancer cell lines and tumors in this study. Moreover, inhibition of miR-1205 aggravated the malignant behaviors of breast cancer cells and partially reversed the effects of circ_0119412 knockdown. Therefore, circ_0119412 may promote breast cancer progression by depleting miR-1205.
GALNT6 plays important roles in carcinogenesis. GALNT6 expression is remarkably high and associated with advanced TNM stage and distant metastasis in patients with gastric cancer [30]. High GALNT6 expression predicts poor prognosis in patients with ovarian cancer [31]. Increased GALNT6 expression has also been reported in breast cancer [32, 33]. Increased expression of GALNT6 is associated with poor prognosis, and its knockdown inhibits the growth of breast cancer cells [32, 33]. Additionally, GALNT6 has been reported to induce breast cancer tumorigenesis and metastasis in vivo [21, 22]. In this study, GALNT6 was found to be a target of miR-1205. Knockdown of GALNT6 repressed proliferation and invasion and enhanced the apoptosis of breast cancer cells. Rescue experiments further revealed that miR-1205 binds to GALNT6 and inhibits its function.
Being a preliminary study exploring the roles of circ_0119412 in breast cancer, this work has some limitations. Whether circ_0119412 overexpression exerts effects antagonistic to those of circ_0119412 downregulation remains unclear and requires further investigation. Moreover, the findings of our study require further validation. Future studies should explore other miRNAs potentially targeted by circ_0119412 to enrich its regulatory networks.
In conclusion, in this study, we demonstrated the abnormally increased expression of circ _0119412 in breast cancer via regulation of the miR-1205/GALNT6 pathway. To the best of our knowledge, this study is the first to reveal the specific roles of circ_0119412 in breast cancer. Our findings provide novel insights into the pathogenesis of breast cancer and suggest targeted inhibition of circ_0119412 as a promising strategy for breast cancer treatment.