Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
GASTROENTEROLOGY / CLINICAL RESEARCH
Antibody-mediated immune responses and inflammatory bowel disease: insights from genetic correlation and Mendelian randomization
1
Center for Diagnosis and Treatment of High Altitude Digestive Diseases, Second People’s Hospital of Xining, Qinghai, China
2
Department of Digestive Diseases, Shaoxing Chinese Medicine Hospital, Zhejiang, China
These authors had equal contribution to this work
Submission date: 2025-02-13
Final revision date: 2025-07-05
Acceptance date: 2025-08-03
Online publication date: 2025-10-26
Corresponding author
Jianzhou Li
Second People’s Hospital of Xining Room 2198, Building 5, No. 37, Wusi St Chengx, 810003, Xining, China
Second People’s Hospital of Xining Room 2198, Building 5, No. 37, Wusi St Chengx, 810003, Xining, China
KEYWORDS
antibody-mediated immune responsesinflammatory bowel diseaselinkage disequilibrium score regressionMendelian randomization
TOPICS
ABSTRACT
Introduction:
Observational studies suggest a complex relationship between infectious diseases and inflammatory bowel disease (IBD), yet the underlying mechanisms remain unclear. This study examined the genetic correlations and causal associations between antibody-mediated immune responses (targeting 46 infectious pathogens) and IBD, using linkage disequilibrium score regression (LDSC) and Mendelian randomization (MR) to inform early prevention and personalized treatment.
Material and methods:
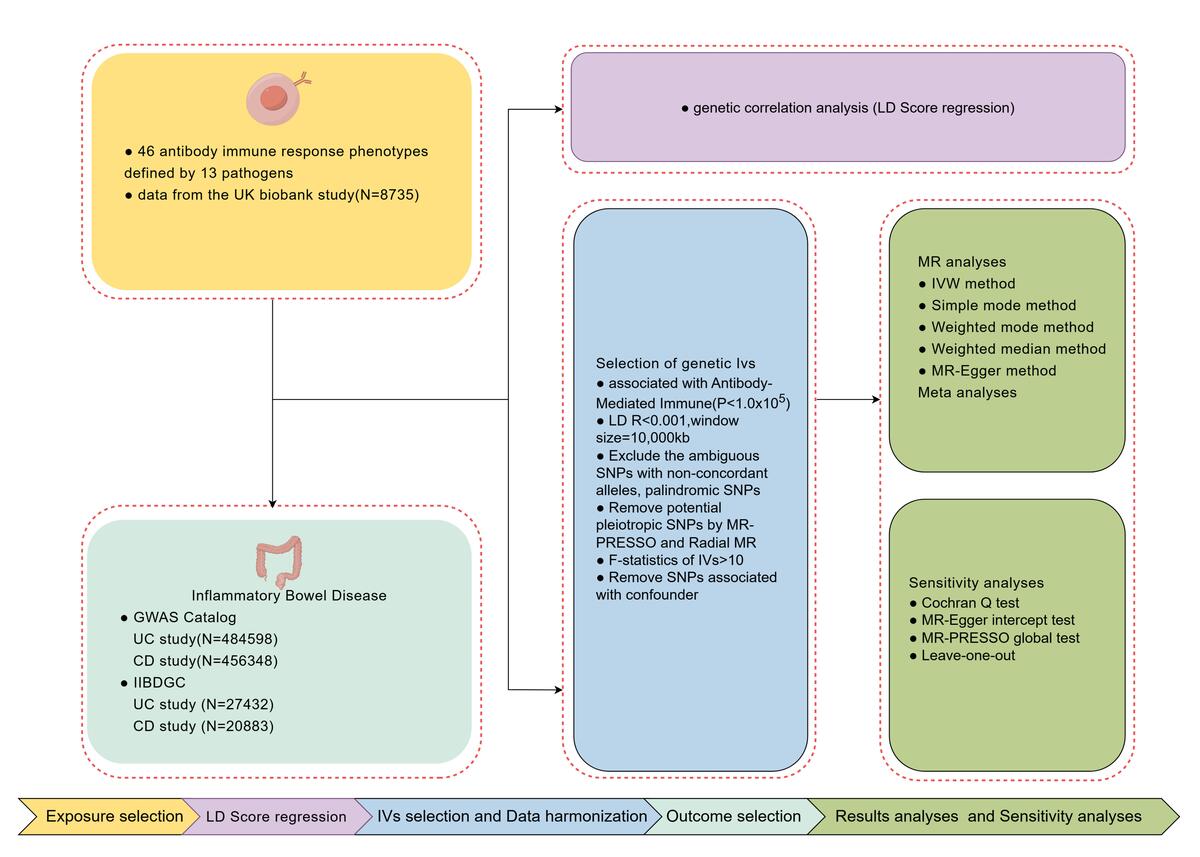
Data from the UK Biobank (8,735 samples) were used to identify independent SNPs associated with antibody responses. IBD-related data from the GWAS Catalog included 2,515 ulcerative colitis (UC) cases and 482,083 controls, as well as 1,342 Crohn’s disease (CD) cases and 455,006 controls. Validation was done using datasets from the International IBD Genetics Consortium (IIBDGC), including 6,968 UC and 5,956 CD cases. Genetic correlations were assessed using LDSC, and causal links were investigated through MR analysis, employing inverse-variance weighting (IVW) and sensitivity analyses.
Results:
Genetic correlations were found between antibody responses to specific pathogens and IBD. MR analysis revealed causal effects of antibody responses to four pathogens on UC and protective/risk effects on CD. No horizontal pleiotropy or heterogeneity was observed, and sensitivity analyses confirmed the consistency of the findings.
Conclusions:
This study provides genetic evidence for causal associations between antibody-mediated immune responses to 46 pathogens and IBD in European populations, offering insights into IBD pathogenesis and implications for prevention and treatment strategies.
Observational studies suggest a complex relationship between infectious diseases and inflammatory bowel disease (IBD), yet the underlying mechanisms remain unclear. This study examined the genetic correlations and causal associations between antibody-mediated immune responses (targeting 46 infectious pathogens) and IBD, using linkage disequilibrium score regression (LDSC) and Mendelian randomization (MR) to inform early prevention and personalized treatment.
Material and methods:
Data from the UK Biobank (8,735 samples) were used to identify independent SNPs associated with antibody responses. IBD-related data from the GWAS Catalog included 2,515 ulcerative colitis (UC) cases and 482,083 controls, as well as 1,342 Crohn’s disease (CD) cases and 455,006 controls. Validation was done using datasets from the International IBD Genetics Consortium (IIBDGC), including 6,968 UC and 5,956 CD cases. Genetic correlations were assessed using LDSC, and causal links were investigated through MR analysis, employing inverse-variance weighting (IVW) and sensitivity analyses.
Results:
Genetic correlations were found between antibody responses to specific pathogens and IBD. MR analysis revealed causal effects of antibody responses to four pathogens on UC and protective/risk effects on CD. No horizontal pleiotropy or heterogeneity was observed, and sensitivity analyses confirmed the consistency of the findings.
Conclusions:
This study provides genetic evidence for causal associations between antibody-mediated immune responses to 46 pathogens and IBD in European populations, offering insights into IBD pathogenesis and implications for prevention and treatment strategies.
REFERENCES (61)
1.
Ramos GP, Papadakis KA. Mechanisms of disease: inflammatory bowel diseases. Mayo Clin Proc 2019; 94: 155-65.
2.
Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol 2011; 17: 557-66.
3.
Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology 2011; 140: 1785-94.
4.
Le Berre C, Danese S, Peyrin-Biroulet L. Can we change the natural course of inflammatory bowel disease? Therap Adv Gastroenterol 2023; 16: 17562848231163118.
5.
Agrawal M, Spencer EA, Colombel JF, Ungaro RC. Approach to the management of recently diagnosed inflammatory bowel disease patients: a user’s guide for adult and pediatric gastroenterologists. Gastroenterology 2021; 161: 47-65.
6.
Alatab S, Sepanlou S, Ikuta K, et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol 2019; 5: 17-30.
7.
Wang R, Li Z, Liu S, Zhang D. Global, regional and national burden of inflammatory bowel disease in 204 countries and territories from 1990 to 2019: a systematic analysis based on the Global Burden of Disease Study 2019. BMJ Open 2023; 13: e065186.
8.
Cho J, Weaver C. The genetics of inflammatory bowel disease. Gastroenterology 2003; 133: 1327-39.
9.
Okazaki T, Wang M-H, Rawsthorne P, et al. Contributions of IBD5, IL23R, ATG16L1, and NOD2 to Crohn’s disease risk in a population-based case-control study: Evidence of gene–gene interactions. Inflamm Bowel Dis 2008; 14: 1528-41.
10.
Xu X-R, Liu C, Feng B, Liu Z. Dysregulation of mucosal immune response in pathogenesis of inflammatory bowel disease. World J Gastroenterol 2014; 20: 3255-64.
11.
Axelrad J, Cadwell K, Colombel J, Shah S. The role of gastrointestinal pathogens in inflammatory bowel disease: a systematic review. Therap Adv Gastroenterol 2021; 14: 17562848211004493.
12.
Lu L, Suscovich T, Fortune S, Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol 2017; 18: 46-61.
13.
Burbelo P, Ching K, Morse C, et al. Altered antibody profiles against common infectious agents in chronic disease. PLoS One 2013; 8: e81635.
14.
Maurano M, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012; 337: 1190-5.
15.
Ferreira M. Ten years of genome-wide association studies of immune-related diseases. Clin Transl Immunol 2018; 7: e1022.
16.
Ning Z, Pawitan Y, Shen X. High-definition likelihood inference of genetic correlations across human complex traits. Nat Genet 2020; 52: 859-64.
17.
Butler-Laporte G, Kreuzer D, Nakanishi T, Harroud A, Forgetta V, Richards JB. Genetic determinants of antibody-mediated immune responses to infectious diseases agents: a genome-wide and HLA association study. Open Forum Infect Dis 2020; 7: ofaa450.
18.
Dönertaş HM, Fabian DK, Valenzuela MF, Partridge L, Thornton JM. Common genetic associations between age-related diseases. Nat Aging 2021; 1: 400-12.
19.
Jiang L, Zheng Z, Fang H, Yang J. A generalized linear mixed model association tool for biobank-scale data. Nat Genet 2021; 53: 1616-21.
20.
Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015; 47: 979-86.
21.
Kurilshikov A, Medina-Gomez C, Bacigalupe R, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet 2021; 53: 156-65.
22.
Li P, Wang H, Guo L, et al. Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med 2022; 20: 443.
23.
Zhong H, Liu S, Zhu J, Wu L. Associations between genetically predicted levels of blood metabolites and pancreatic cancer risk. Int J Cancer 2023; 153: 103-10.
24.
Liu X, Tong X, Zou Y, et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat Genet 2022; 54: 52-61.
25.
Staley JR, Blackshaw J, Kamat MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 2016; 32: 3207-9.
26.
Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019; 35: 4851-3.
27.
Staiger DO, Stock JH. Instrumental variables regression with weak instruments. National Bureau of Economic Research Cambridge, Mass., USA, 1994.
28.
Pierce BL, Ahsan H, VanderWeele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 2011; 40: 740-52.
29.
Van Rheenen W, Peyrot W, Schork A, Lee S, Wray N. Genetic correlations of polygenic disease traits: from theory to practice. Nat Rev Genet 2019; 20: 567-81.
30.
Li T, Ning Z, Shen X. Improved estimation of phenotypic correlations using summary association statistics. Front Genet 2020; 12: 665252.
31.
Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47: 291-5.
32.
Pettit R, Amos C. Multiple outcome linkage disequilibrium score regression for confounding independent genetic correlations. Cancer Res 2022; 8 (12 Suppl.): 5041.
33.
Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 2017; 46: 1985-98.
34.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013; 37: 658-65.
35.
Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018; 50: 693-8.
36.
Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018; 7: e34408.
37.
Corbin LJ, Richmond RC, Wade KH, et al. BMI as a modifiable risk factor for type 2 diabetes: refining and understanding causal estimates using Mendelian randomization. Diabetes 2016; 65: 3002-7.
38.
Bowden J, Spiller W, Del Greco MF, et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol 2018; 47: 1264-78.
39.
Vasilenko N, Tieck MP, Michel T, et al. In-depth analysis of serum antibodies against Epstein-Barr virus lifecycle proteins, and EBNA1, ANO2, GlialCAM and CRYAB peptides in patients with multiple sclerosis. Front Immunol 2024; 15: 1487523.
40.
Lim CS, Goh SL, Kariapper L, Krishnan G, Lim YY, Ng CC. Inclusion bodies of recombinant Epstein-Barr virus capsid antigen p18 as potential immobilized antigens in enzyme immunoassays for detection of nasopharyngeal carcinoma. Clin Chim Acta 2015; 448: 206-10.
41.
Vietzen H, Berger SM, Kühner LM, et al. Ineffective control of Epstein-Barr-virus-induced autoimmunity increases the risk for multiple sclerosis. Cell 2023; 186: 5705-18.e13.
42.
Soldan S, Su C, Monaco MC, et al. Unstable EBV latency drives inflammation in multiple sclerosis patient derived spontaneous B cells. Res Sq 2023; rs.3.rs-2398872.
43.
Wang W, Chen X, Pan J, Zhang X, Zhang L. Epstein-Barr virus and human cytomegalovirus infection in intestinal mucosa of Chinese patients with inflammatory bowel disease. Front Microbiol 2022; 13: 915453.
44.
Sankaran-Walters S, Ransibrahmanakul K, Grishina I, et al. Epstein-Barr virus replication linked to B cell proliferation in inflamed areas of colonic mucosa of patients with inflammatory bowel disease. J Clin Virol 2011; 50: 31-6.
45.
Zhang H, Zhao S, Cao Z. Impact of Epstein-Barr virus infection in patients with inflammatory bowel disease. Front Immunol 2022; 13: 1001055.
46.
Santarelli R, Evangelista L, Pompili C, et al. EBV infection of primary colonic epithelial cells causes inflammation, DDR and autophagy dysregulation, effects that may predispose to IBD and carcinogenesis. Virus Res 2023; 338: 199236.
47.
Guerra G, McCoy L, Hansen HM, et al. Antibodies to varicella-zoster virus and three other herpesviruses and survival in adults with glioma. Neuro Oncol 2023; 25: 1047-57.
48.
Seitel T, Cagol L, Prelog M, et al. Varicella-zoster-virus vaccination of immunosuppressed children with inflammatory bowel disease or autoimmune hepatitis: a prospective observational study. Vaccine 2020; 38: 8024-31.
49.
Ra SH, Kwon JS, Kim JY, et al. Frequency of putative enteric zoster diagnosed using saliva samples in patients with abdominal pain: a prospective study. Infect Dis (Lond) 2021; 53: 713-8.
50.
Levin MJ, Ginde AA, Schmid DS, et al. Effect of high dose vitamin D supplementation on subsequent immune responses to administration of the live herpes zoster vaccine to long-term care residents. Vaccine 2024; 42: 2278-81.
51.
Antonucci L, Locci C, Schettini L, Clemente MG, Antonucci R. Infant botulism: an underestimated threat. Infect Dis (Lond) 2021; 53: 647-60.
52.
Eberhardt CS, Wieland A, Nasti TH, et al. Persistence of varicella-zoster virus-specific plasma cells in adult human bone marrow following childhood vaccination. J Virol 2020; 94: e02127-19.
53.
Marashi SM, Raeiszadeh M, Workman S, et al. Inflammation in common variable immunodeficiency is associated with a distinct CD8(+) response to cytomegalovirus. J Allergy Clin Immunol 2011; 127: 1385-93.e4.
54.
Contreras NA, Sitnik KM, Jeftic I, Coplen CP, Čičin-Šain L, Nikolich-Žugich J. Life-long control of cytomegalovirus (CMV) by T resident memory cells in the adipose tissue results in inflammation and hyperglycemia. PLoS Pathog 2019; 15: e1007890.
55.
Wang S, Dou Y, Yang H, Ni A, Zhang R, Qian J. Alteration of glucocorticoid receptors and exacerbation of inflammation during lytic cytomegalovirus infection in THP-1 cells. FEBS Open Bio 2017; 7: 1924-31.
56.
Jentzer A, Veyrard P, Roblin X, et al. Cytomegalovirus and inflammatory bowel diseases (IBD) with a special focus on the link with ulcerative colitis (UC). Microorganisms 2020; 8: 1078.
57.
Di Benedetto S, Gaetjen M, Müller L. The modulatory effect of gender and cytomegalovirus-seropositivity on circulating inflammatory factors and cognitive performance in elderly individuals. Int J Mol Sci 2019; 20: 990.
58.
Miura M, Shimizu H, Saito D, et al. Multicenter, cross-sectional, observational study on Epstein-Barr viral infection status and thiopurine use by age group in patients with inflammatory bowel disease in Japan (EBISU study). J Gastroenterol 2021; 56: 1080-91.
59.
Subramaniam K, Cherian M, Jain S, et al. Two rare cases of Epstein-Barr virus-associated lymphoproliferative disorders in inflammatory bowel disease patients on thiopurines and other immunosuppressive medications. Intern Med J 2013; 43: 1339-42.
60.
Gracienta TJ, Herardi R, Santosa F, Pasiak TF, Tjang YS. Diagnostic accuracy of antibody-based rapid diagnostic tests in detecting coronavirus disease 2019: systematic review. Arch Med Sci 2022; 18: 949-57.
61.
Zdanowicz K, Pietrowska K, Lebensztejn DM, et al. Evaluation of microbiome composition combined with serum untargeted metabolomic profiling in newly diagnosed children with inflammatory bowel disease. Arch Med Sci 2025; 21: 416-24.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.