Introduction
Chronic obstructive pulmonary disease (COPD) is the third most common cause of global mortality, affecting over 210 million individuals [1]. Despite extensive research efforts, molecular mechanisms underlying COPD remain poorly understood. Tobacco smoking is the major risk factor for developing COPD, but not all smokers will develop the disease. This suggests that, in addition to tobacco smoking, COPD is also influenced by genetic risk factors. Understanding the genetic predisposition to COPD has been the aim of many investigators during the past decade, since it is essential to develop more effective COPD prevention strategies.
Matrix metalloproteinases (MMPs) are a family of structurally related, zinc-dependent proteinases that degrade extracellular matrix (ECM), cell surface, and intracellular proteins [2]. They are the major proteinases responsible for the digestion and degradation of structural proteins of the lung. There is significant evidence that MMPs may play a critical role in COPD pathology. Increased levels of MMP-3, MMP-9 and MMP-12 have been observed in patients with COPD as well as smoke exposed animals [3–6]. In contrast to wild-type mice, mice lacking MMP-12 did not have increased numbers of macrophages in their lungs and did not develop emphysema after chronic cigarette smoke exposure [7]. Similarly, genetic blockade of MMP-9 in mice significantly attenuated emphysema pathology [8]. Since MMP activity can be regulated at the level of transcription, functional MMP polymorphisms may contribute to COPD development by modifying MMP gene expression. Several MMP genetic polymorphisms including MMP-3 rs35068180, MMP-9 rs3918242, MMP-12 rs2276109, and MMP-12 rs652438 have been extensively evaluated for their associations with COPD risk in different populations; the number of published articles on the subject has grown substantially over the past several years. However, a major limitation in the extant literature is that many studies lack sufficient statistical power due to relatively small sample sizes. Using published data from various countries and cohorts, we provide a comprehensive meta-analysis to evaluate the association of polymorphisms in the MMP-3, MMP-9, and MMP-12 genes with COPD risk.
Material and methods
Search strategy
We systematically searched PubMed, Web of Knowledge, China National Knowledge Infrastructure (CNKI), and Wanfang databases from their commencements until October 2019. The search was performed using the following keywords and subject terms: matrix metalloproteinase, chronic obstructive pulmonary disease, emphysema, polymorphism, case-control, and association. In addition, previous review papers and meta-analyses were used to cross-reference and trace back to all original studies.
Inclusion criteria
Two reviewers screened the search results to select studies for possible inclusion. Discrepancies regarding eligibility were resolved by discussion. The eligibility criteria included the following: (1) original study; (2) use of human subjects; (3) case-control design; and (4) evaluation of the association between MMP genetic polymorphisms and COPD risk. Studies lacking genotypes for case and controls or appropriate genotype allele counts were excluded. The quality of each study was evaluated with the Newcastle-Ottawa Quality Assessment Scale (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp), which assesses each study’s selection, comparability, and exposure ascertainment approach.
Data extraction
Data extraction was performed by two reviewers using standard data extraction forms. Any data discrepancy was resolved by referring back to the original papers. Study characteristics that were abstracted included publication year, first author, region or country of study, diagnostic criteria for COPD, COPD stage, sample size, number of COPD cases, number of controls, genotype frequency, inclusion age and range, male percentage, smoking history, and genotyping method. Ethnicity was classified as Caucasian (European, Arabian, Turkish and Iranian), Asian (Chinese, Korean, and Japanese) and mixed (American, Brazilian and Mexican) [9]. Supplemental material was also checked for the identification and extraction of relevant data.
Statistical analysis
This meta-analysis was reported in accordance with the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) checklist [10]. The pooled ORs of MMP-3 rs35068180, MMP-9 rs3918242, MMP-12 rs2276109, and MMP-12 rs652438 were estimated using both fixed and DerSimonian and Laird random-effect models. Statistical heterogeneity was evaluated by χ2 and I2 statistics. An I2 value < 25% was considered to represent low heterogeneity and > 75% as high heterogeneity. We used an exact test to evaluate whether the genotype frequencies in control groups were in Hardy-Weinberg equilibrium (HWE). A sensitivity analysis was conducted to assess the influence of the individual studies on the overall results by omitting one study at a time. Funnel plots in conjunction with Begg’s test were undertaken to assess publication bias. All meta-analysis calculations and plots were done using Stata 12.0.
Results
Description of included studies
The primary search of electronic databases retrieved 255 citations. Articles were screened based on title and abstracts, and 28 articles were found initially eligible for full-text review. Ultimately, 23 met the predefined inclusion criteria and were included in our meta-analysis [11–33]. Details of the search process are presented in Figure 1. Studies involved participants from five continents including Asia, Europe, North America, South America and Africa. Sample sizes varied between 112 and 1853 participants. All studies were conducted in adult populations. The methods of genotyping included polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), allelic discrimination assay, sequencing, and competitive allele specific PCR (KASPar) assay. Quality analysis showed that most of the included studies were rated as being of high or moderately high quality. The characteristics of the included studies are reported in Tables I–IV. Forest plots depicting effect sizes and their CIs for the included studies are presented in Figures 2 and 3.
Figure 2
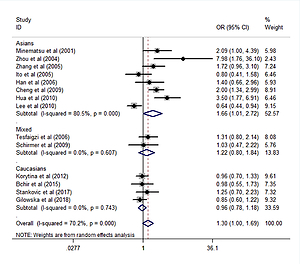
Forest plots for meta-analysis of the MMP-9 rs3918242 polymorphism (T/T + C/T vs. C/C) and COPD risk
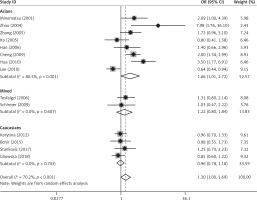
Figure 3
Forest plots for meta-analysis of the MMP-12 rs2276109 polymorphism (G/G + A/G vs. A/A) and COPD risk
Table I
Summary characteristics of studies investigating the MMP-9 rs3918242 polymorphism
Table II
Summary characteristics of studies investigating the MMP-12 rs2276109 polymorphism
Table III
Summary characteristics of studies investigating the MMP-12 rs652438 polymorphism
Table IV
Summary characteristics of studies investigating the MMP-3 rs35068180 polymorphism
Meta-analysis of MMP polymorphisms and COPD risk
The MMP-9 rs3918242 polymorphism was evaluated in 14 studies with a total of 4733 subjects (2287 cases and 2446 controls) [11, 13–17, 19, 20, 22, 23, 25, 29, 30, 32]. Genotypes in each of these studies were in HWE. Pooling the data from these studies showed no association between the polymorphism and COPD risk under the dominant (T/T + C/T vs. C/C: OR = 1.30, 95% CI: 1.00–1.69, p = 0.054) and allele contrast (T allele vs. C allele: OR = 1.22, 95% CI: 0.97–1.53, p = 0.088) models (Table V and Figure 2). However, in ethnicity subgroup analysis, there was a significant association in the dominant model (T/T + C/T vs. C/C: OR = 1.66, 95% CI: 1.02–2.72, p = 0.043) among Asian subjects (Table V, Figure 2). No association between the polymorphism and COPD risk was found in Caucasians and mixed ethnic groups (Table V, Figure 2). There was no statistically significant evidence for heterogeneity among Caucasians and mixed ethnic groups (p > 0.10), but heterogeneity was substantial in Asians (p < 0.10) (Table V).
Table V
Meta-analysis of MMP polymorphisms and COPD risk
Nine studies involving 2654 cases and 3834 controls reported data on the association between the MMP-12 rs2276109 polymorphism and COPD risk [12, 14, 19, 21, 24, 25, 28, 30, 33]. No deviations from HWE were detected. Significant heterogeneity existed between the studies (p < 0.001). Meta-analysis using the random effects model did not identify a significant association in the overall population under the dominant (G/G + A/G vs. A/A: OR = 0.94, 95% CI: 0.72–1.24, p = 0.674) and allele contrast (G allele vs. A allele: OR = 0.94, 95% CI: 0.75–1.18, p = 0.580) models (Table V, Figure 3). When stratifying the results of meta-analysis by ethnicity, we found that rs2276109 was associated with increased risk of COPD in mixed populations under the dominant (G/G + A/G vs. A/A: OR = 1.57, 95% CI: 1.10–2.24, p = 0.013) and allele contrast (G allele vs. A allele: OR = 1.52, 95% CI: 1.09–2.14, p = 0.015) models (Table V, Figure 3). No significant association was observed in Asians and Caucasians (Table V, Figure 3).
Five studies in 4217 subjects (1685 cases and 2532 controls) assessed the association between the MMP-12 rs652438 polymorphism and COPD risk [14, 21, 22, 24, 27]. No significant association was observed between the polymorphism and COPD in the overall population under the dominant (G/G + A/G vs. A/A: OR = 0.71, 95% CI: 0.47–1.08, p = 0.113) and allele contrast (G allele vs. A allele: OR = 0.72, 95% CI: 0.47–1.09, p = 0.121) models (Table V). After exclusion of the studies whose controls significantly departed from HWE [14, 27], the results remained unchanged (Table V). Stratified analysis by ethnicity did not suggest any association between rs652438 and COPD risk in Caucasians (Table V). Since two Asian studies deviated from HWE [14, 27], stratified analysis was not performed for Asians.
Six studies reported data on the relationship between the MMP-3 rs35068180 polymorphism and COPD risk [18, 19, 25–27, 31]. In the overall meta-analysis, rs35068180 was not associated with COPD risk under the dominant (5A/5A + 5A/6A vs. 6A/6A: OR = 0.92, 95% CI: 0.62–1.36, p = 0.679) and allele contrast (5A allele vs. 6A allele: OR = 0.92, 95% CI: 0.70–1.21, p = 0.557) models. The distribution of genotypes in the control group deviated from HWE in one study [26]. Removal of this study did not significantly influence the pooled effect sizes (5A/5A + 5A/6A vs. 6A/6A: OR = 0.96, 95% CI: 0.60–1.54, p = 0.878; 5A allele vs. 6A allele: OR = 0.94, 95% CI: 0.68–1.29, p = 0.557) (Table V).
Publication bias
Publication bias was evaluated by performing Begg’s test under the dominant and allele contrast models. The results provided no statistical evidence of publication bias (Table VI).
Discussion
In this meta-analysis, we evaluated the relationship of the MMP-3, MMP-9, and MMP-12 polymorphisms with COPD risk. Our results indicated that the MMP-9 rs3918242 polymorphism was associated with increased risk of COPD in Asians, whereas the MMP-12 rs2276109 polymorphism contributed to increased COPD risk in mixed populations. We did not find any significant association of the MMP-3 rs35068180 and MMP-12 rs652438 polymorphisms with COPD risk.
MMP-9, also known as gelatinase B, is a type IV collagen-degrading enzyme that is produced in large quantities by inflammatory cells. A number of studies have shown that MMP-9 levels were elevated in the airways and blood of COPD patients compared with control individuals [4, 6, 34]. Moreover, elevated plasma MMP-9 was associated with distinct clinical features and the risk of acute exacerbations in COPD populations, suggesting that MMP-9 was a potential biomarker to monitor COPD progression [35, 36]. There results were supported by findings from experimental studies. Genetic MMP-9 deletion reduced cigarette smoke induced emphysema in mice [37], whereas transgenic expression of MMP-9 caused adult-onset emphysema [38]. MMP-9 may contribute to COPD pathogenesis through several possible mechanisms, including extracellular matrix degradation, modulation of airspace enlargement, neutrophil chemotaxis, activation of proinflammatory cytokines, and augmentation of pulmonary inflammation [6, 37]. The MMP-9 rs3918242 polymorphism is a common cytosine-to-thymidine mutation at position -1562 within the MMP-9 gene promoter [11]. Previous studies have shown that the T allele of this SNP was associated with increased promoter activity and higher MMP-9 levels [14, 23]. Given the prominent role of MMP-9 in COPD pathogenesis, dysregulation of MMP-9 expression induced by this SNP may increase the risk of COPD in genotypes containing the T allele. This meta-analysis demonstrated that the MMP-9 rs3918242 polymorphism was associated with increased risk of COPD in Asians but not in other ethnic groups. Ethnic differences regarding this polymorphism have also been found in other diseases such as stroke and coronary heart disease (CHD); rs3918242 was associated with stroke and CHD risk in Asians but not in Caucasians [39, 40]. Clinical heterogeneity, small sample size, genetic background, and different patterns of gene-gene and gene-environment interactions may be the main reasons for ethnic difference.
MMP-12 (matrix metalloelastase) is another metalloproteinase involved in airway inflammation and tissue remodeling processes in COPD. Inflammatory macrophages are the major sources of MMP-12. Upregulation of MMP-12 was observed in patients with COPD as well as smoking-induced emphysema [3, 7, 25]. In animal studies, inhibition of MMP-12 substantially ameliorated morphological emphysema, small airway remodeling and the functional consequences of these lesions, suggesting that MMP-12 may represent a therapeutic target for COPD [41]. These lines of evidence also suggested that high-level expression of MMP-12 may increase the individual predisposition to COPD. Our meta-analyses indicated that the MMP-12 rs2276109 polymorphism (-82A/G) was associated with increased risk of COPD in mixed populations. A possible explanation for this finding is that the MMP-12 rs2276109 polymorphism is a functional variant resulting in a higher affinity for the transcription factor activator protein-1 and increased MMP-12 expression [25]. However, considering the small number of included studies, further investigation using larger cohorts should be performed to verify our results.
The MMP-3 rs35068180 polymorphism is a common adenine insertion/deletion variant at position 1171 of the MMP-3 gene promoter affecting transcription factor binding and promoter activity [18], whereas the MMP-12 rs652438 polymorphism is an asparagine to serine substitution at codon 357 of the MMP-12 gene that is associated with a greater binding affinity for activator protein 1 [14]. Our meta-analysis did not find significant associations of these polymorphisms with COPD risk.
Although two previous meta-analyses focused on the topic, there were some limitations [42, 43]. First, erroneous inclusion and exclusion criteria were used. For assessing the relationship of the MMP-3 rs35068180 polymorphism (-1171 5A>6A) with COPD, both the Cheng et al. study and the Jiang et al. study mistakenly included a case-control study investigating the MMP-3 rs3025058 polymorphism (-1612 5A>6A) [20]. Second, when evaluating the association of the MMP-9 rs3918242 and MMP-12 rs2276109 polymorphisms with COPD risk, a Russian sample was repeatedly used to calculate the pooled effect sizes [42]. It is widely accepted that when the same sample is used in several studies, only the novel data should be included in a meta-analysis. Third, Jiang et al. did not check for HWE in control subjects among all publications [43]. In fact, several case-control studies deviating from HWE were included in their meta-analyses. Therefore, it may be necessary to perform a sensitivity analysis to ensure the robustness of the results.
For the MMP-9 rs3918242 polymorphism, Zhou et al. found no association of this polymorphism with COPD risk in the overall meta-analysis, but they reported an association between it and COPD in stratified analysis by subgroup variables such as genotyping method and matching strategies. The study by Jiang et al. demonstrated a significant association between the MMP-9 rs3918242 polymorphism and risk of COPD in the overall meta-analysis, but they did not perform any subgroup analyses. Both of these meta-analyses suggested that the MMP-9 rs3918242 polymorphism may be a genetic risk factor of COPD. Our results supported their findings, but we found that the MMP-9 rs3918242 polymorphism was associated with COPD risk only among Asians.
Our study has several strengths. First, we used a more comprehensive search strategy and identified additional studies which were not included in the previous meta-analyses. Our study is the most comprehensive meta-analysis evaluating the relationship of the MMP-3, MMP-9 and MMP-12 polymorphisms with COPD risk to date. Second, we evaluated the association between the common MMP-12 polymorphism rs652438 (Asp357Ser) and COPD risk, which was never taken into account by the previous meta-analyses. Third, meta-analyses regarding the association of the MMP-3 rs35068180 and MMP-12 rs652438 polymorphisms with COPD were systematically conducted before and after the exclusion of the studies deviating from HWE. Fourth, the methodological quality of the included studies was evaluated before performing the meta-analyses.
The present study has several shortcomings that should be acknowledged. First, we included case-control studies involving small samples with limited statistical power. The small number of studies assessing the MMP-3 rs35068180 and MMP-12 rs652438 polymorphisms limited the number of stratified analyses that could be performed. More research on the topic is needed. Second, owing to insufficient individual information on genotypes, we were unable to conduct a linkage disequilibrium analysis to evaluate the possible interactions between MMP polymorphisms. Third, due to the limited data provided by the studies, an evaluation of the relationship of the MMP polymorphisms with smoking and lung function in COPD patients could not be performed.
In conclusion, the present study provides a comprehensive review of the literature on the association between four common MMP polymorphisms and COPD risk. The results of the meta-analysis indicate that the MMP-9 rs3918242 polymorphism is associated with increased risk of COPD in Asians, and the MMP-12 rs2276109 polymorphism is associated with increased COPD risk in mixed populations.