Introduction
Inflammatory bowel disease (IBD) is a chronic inflammatory bowel disease that mainly affects the digestive system, among which Crohn’s disease (CD) and ulcerative colitis (UC) are the main types [1]. In recent decades, with the industrialization and urbanization of non-developed countries and regions such as Asia, South America, and the Middle East, IBD has begun to appear in newly industrialized countries. At present, IBD has become a global disease. According to the Lancet Gastroenterol Hepatol statistical report, as of 2017, there were 6.8 million (95% UI: 6.4–7.3) IBD cases worldwide [2]. The age-standardized prevalence rate increased from 79.5 per 100,000 population in 1990 to 84.3 per 100,000 population in 2017. As a chronic inflammatory intestinal disease, IBD mainly affects the digestive system, but also invades organs and tissues outside the intestinal tract, which ultimately harms human health and affects the quality of life of patients. However, at present, the etiology and the pathogenesis of IBD are still unclear. Environment, diet, and microorganisms tend to act on genetically susceptible hosts to trigger abnormal immune responses, resulting in chronic intestinal inflammation. The degree of attention to it at home and abroad and the research efforts of scientific research institutions are also constantly strengthening and deepening.
Cardiovascular disease (CVD) is the main cause of global mortality and disability, and it has also become an important public health problem worldwide. According to the Global Burden of Disease research data [3], as of 2017, about 17.79 million people worldwide had died of CVDs, accounting for 31.8% of all global deaths. Therefore, it is very important to identify the risk factors and protective factors of CVDs in daily life. In recent years, the relationship between IBD and CVD has attracted the attention of researchers. Epidemiological studies [4] have demonstrated that patients with IBD are more likely to suffer from CVD than the general population, and the severity of IBD is positively correlated with the risk of CVD. However, so far, no exact causal relationship between IBD and CVDs has been confirmed, and more research is needed to clarify this relationship.
In Mendelian randomization (MR) analysis, single nucleotide polymorphisms (SNPs) which have a strong relationship with exposure, for example IBD, are considered as instrumental variables (IVs) to estimate the causal effect with outcome (e.g., CVDs). MR is a ‘natural’ RCT that makes use of the random distribution of genetic variants with an influence on exposure [5–7]. Those SNPs that are strongly linked with confounders will be eliminated before performing the MR analysis in order to remove the effect of confounding factors. Reverse MR analysis can exclude potential reverse causal effect between exposures and outcomes [8–10].
Therefore, in order to further explore whether there is an exact association between IBD and the risk of CVDs, we conducted an MR study, using large-scale publicly available genome-wide association study (GWAS) data, using two disease types of IBD, CD and UC, as exposure factors, and CVDs as a result, to evaluate the causal relationship between IBD and CVD risk.
Material and methods
Study design
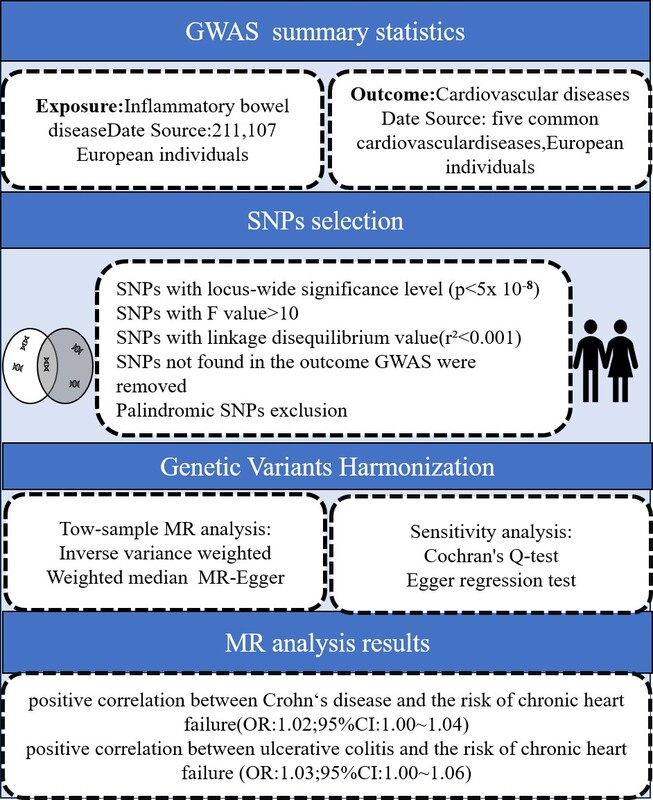
A two-sample MR study was designed to evaluate the potential causal relationship between IBD and CVDs. The SNPs were selected as IVs according to three essential assumptions: (1) SNPs should be strongly associated with IBD as the exposure; (2) SNPs should not be associated with confounding factors; and (3) SNPs should not be directly associated with CVDs as the outcome (Figure 1).
GWAS summary statistics
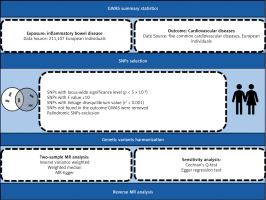
SNPs was used as a tool variable in this study, and the data sources were all publicly available GWAS databases. The classification of the exposure factor IBD includes two subtypes – CD and UC – and SNPs are all from the FinnGen genome-wide association study sample database. The CD samples included 210,300 controls and 807 cases, and the UC samples included 215,806 controls and 2701 cases. The samples included are all European samples. Data on atrial fibrillation were obtained from the public GWAS database compiled by the European Institute of Bioinformatics (EBI) database, including 60,620 cases and 970,216 controls. Coronary heart disease data (60,801 cases and 123,504 controls) and myocardial infarction data (43,676 cases and 128,199 controls) were derived from a meta-analysis of coronary artery disease. Data on chronic heart failure were obtained from 26 studies conducted by the global HERMES (HEart failure Molecular Epidemiology for Therapeutic targetS) coalition, including 47,309 cases and 930,014 controls. Hypertension data (54,358 cases and 408,652 controls) were collected from the UK Biobank. The above samples come from different databases, and the possibility of sample overlap is very small. The study was conducted in strict accordance with the reporting guidelines for Mendelian randomized studies. All the studies were conducted in accordance with the Declaration of Helsinki and approved by the appropriate institutional ethics committees, so no additional ethical approval is required. All the raw data in this study can be obtained from the IEU Open GWAS database (http://gwas.mrcieu.ac.uk). Table I presents details of the exposure and outcomes.
Table I
Detailed information on exposure and outcomes
SNP selection
Firstly, we conducted a screening process to identify SNPs that were highly correlated with exposure at a genome-wide significance level (p < 5 × 10–8). Secondly, we implemented a criterion (r2 < 0.001, kb = 10,000) to choose SNPs that were free from dependence on linkage disequilibrium (LD). Thirdly, we excluded SNPs that were not present in the IBD dataset and palindromic SNPs which have the potential to introduce bias. Subsequently, we ensured the harmonization of exposure and outcome data, confirming that the effect of the SNP on the exposure corresponded to the same allele as its effect on the outcome. Following this, we assessed the possibility of weak instrumental bias by calculating F-statistics, and excluded SNPs with F-statistics less than 10. The F statistic was calculated using the formula F = β2/SE2. Finally, we employed the MR-PRESSO method to identify outlier SNPs. After removing the outliers, the remaining SNPs were utilized for subsequent MR analysis. A flowchart illustrating the selection process is provided in Figure 2.
Two-sample Mendelian analysis
In this study, IVW was used to evaluate the potential causal relationship between inflammatory bowel disease and the risk of CVD by calculating the OR and 95%CI. MR-Egger regression, weighted median (WME), and weighted mode (WM) were used as supplementary methods of IVW. When the IVW method yielded p < 0.05 and MR-Egger regression, WME and WM had the same direction, it was considered a relatively stable causal correlation [11].
Statistical analysis
In addition, the recognizable horizontal multiplicity of the MR-Egger intercept term (Egger-intercept) was analyzed [12–14]. If the Egger-intercept was close to 0 or there was no statistical significance, it indicated that there was no genetic pleiotropy [14]. The MR-PRESSO method, which could detect possible outliers and re-conduct causal analysis after excluding outliers, was also used in this study. The sensitivity analysis was carried out by using the leave-one-out method, and the combined effect value of the remaining SNPs was calculated by deleting a single SNP in turn, so as to evaluate the influence of each SNP on causality. In order to measure the degree of heterogeneity, Cochran’s Q test and IVW statistics were used to quantify the heterogeneity of IVs and to evaluate whether there was potential heterogeneity among IVs [15, 16].
Reverse MR analysis
In order to evaluate the causal relationship between inflammatory bowel disease and CVD, this study also conducted a reverse MR analysis of the exposure factors found to be causally related to CVD in forward MR analysis. The methods and settings used were consistent with those used in the forward analysis.
All statistical analyses were conducted using R software (version 4.2.3) with the “Two Sample MR” (version 0.5.6), “MRPRESSO” (version 1.0), and “Mendelian Randomization” (version 0.7.0) packages.
Results
According to the set screening criteria, we screened out four CD-related and independent SNPs, 12 related and independent SNPs from the IEUGWAS database, and the F value of each SNP was greater than 10, indicating that there were no weak tool variables.
MR analysis results
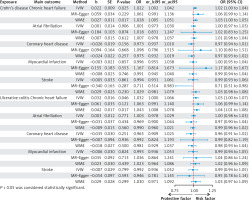
We observed a positive correlation between genetic prediction of CD and the risk of chronic heart failure (IVW (OR = 1.02; 95% CI: 1.00–1.04; p = 0.025)); there was a positive correlation between UC and the risk of chronic heart failure (IVW (OR = 1.03; 95% CI: 1.00–1.06; p = 0.04)); no causal association was observed between CD and UC and atrial fibrillation, coronary heart disease, myocardial infarction or hypertension.
MR-Egger regression analysis of CD and chronic heart failure yielded OR = 1.06; 95% CI: 0.99–1.36; p = 0.22; WME analysis yielded OR = 1.02; 95% CI: 1.00–1.05; p = 0.01. MR-Egger regression analysis of UC and chronic heart failure yielded OR = 1.06; 95% CI: 0.99–1.14; p = 0.12; WME analysis yielded OR = 1.04; 95% CI: 1.00–1.07; p = 0.01. Detailed results of the analysis are shown in Figure 3.
Results of sensitivity analysis
The heterogeneity test of CD and chronic heart failure by the IVW method revealed no heterogeneity in IVW method, and there was no heterogeneity in the horizontal multiplicity test. The heterogeneity test between UC and chronic heart failure showed no significant heterogeneity: IVW method, p = 0.17, MR-Egger regression test, p = 0.17, no heterogeneity; horizontal pleiotropy test, p = 0.37, no horizontal multiplicity. MR-PRESSO global inspection found no outliers. The F statistics of all SNP are greater than 10, indicating that the results of MR analysis are not affected by weak IV bias. The remaining one method did not find any SNP loci in the tool variables that had a strong influence on the results, which indicated that the results were robust. Detailed data are shown in Table II.
Table II
Results of sensitivity analysis of IBD and CVDs
Discussion
Inflammatory bowel disease (IBD) is a chronic inflammatory bowel disease with complex etiology, and its risk factors are still unclear [17]. In recent years, Mendelian randomization (MR) has received more and more attention in IBD research [17–19]. MR mainly uses genetic variation as a tool to derive the causal relationship between exposure and outcome, which effectively avoids the influence of confounding factors and reverse causality in observational studies and randomized controlled trials. In this study, MR analysis was used to evaluate the causal relationship between IBD and CVDs in the European population. Based on the large-scale GWAS database, it was found that there was a positive correlation between CD, UC, and chronic heart failure, but no causal correlation between IBD and other CVDs.
The conclusions regarding the relationship between IBD and CVDs are not consistent in related studies. A national cohort study from Denmark [20] found that disease activity in IBD was associated with an increased risk of myocardial infarction and stroke. The study also confirmed that patients with CD had a higher risk of stroke than UC. A review [21] showed that CD and UC are associated with an increased risk of coronary heart disease, stroke, and atrial fibrillation. UC is also associated with an increased risk of heart failure. Several other clinical studies and reviews have reached similar conclusions [22–24]. However, a series of MR analyses have drawn different conclusions: for example, MR analysis by Qiu et al. found no causal relationship between genetically predicted IBD and coronary heart disease risk in people of European descent [25]. The MR analysis of Chen et al. also does not support the causal relationship between IBD and atrial fibrillation [26]. Wu et al. assessed the causal effects of IBD and different CVD phenotypic risks through MR analysis, and also concluded that there was no clear causal relationship between IBD and a variety of CVD outcomes [27]. In this study, two-sample MR analysis was used to draw the conclusion that there was a positive correlation between UD and UC and chronic heart failure, which was not consistent with the results of previous MR studies, but confirmed the conclusions of relevant clinical cohort studies and retrospective analysis. The reason may be that CVD is a highly complex disease phenotype, which is determined by congenital genetic and environmental factors, and cannot explain all the phenotypic variation from a genetic point of view.
The observed association between IBD and CHF may be mediated by chronic systemic inflammation, a hallmark of IBD. Proinflammatory cytokines (e.g., TNF-α, IL-6) promote endothelial dysfunction, myocardial fibrosis, and adverse cardiac remodeling. Persistent inflammation may also accelerate atherosclerosis, impair ventricular compliance, and reduce coronary microvascular perfusion. Furthermore, IBD-related comorbidities, such as anemia, malnutrition, and corticosteroid use, could exacerbate cardiac strain. Autoimmune cross-reactivity between gut and cardiac tissues has been hypothesized but requires further validation.
Our findings suggest that IBD patients, particularly those with active disease, may benefit from routine cardiovascular risk assessments. While universal screening for LVEF deterioration in all IBD patients is not yet supported by current evidence, targeted monitoring should be considered for high-risk subgroups (e.g., elderly patients, those with prolonged disease duration, or elevated systemic inflammation). Biomarkers such as BNP (B-type natriuretic peptide), NT-proBNP, and high-sensitivity troponin could be integrated into clinical practice to identify early cardiac dysfunction. Additionally, inflammatory markers such as C-reactive protein (CRP) and fecal calprotectin may help stratify patients with heightened cardiovascular risk.
The advantages of this study are as follows: 1) This study used an MR analysis method to explore the causal relationship between IBD and CVDs, which reduced the possibility of outcome bias caused by related residual factors, confounding factors, or reverse causality to the greatest extent [28]. 2) Through several sensitivity analyses to meet the core assumptions of MR, we identified genetic variation which is strongly related to phenotype. We independently excluded genetic variation related to potential confounding factors, and ensured the accuracy of the results. 3) MR studies are affected by multiple factors, and this study used MR-Egger regression to ensure the robustness of the results. 4) All the data used in this study are available free of charge online.
There are still some limitations of this study: 1) All the GWAS data selected are from the European population, so whether the conclusions of this study are applicable to other populations remains to be further tested. 2) Genetic factors alone cannot explain all phenotypic variations, and environmental factors (e.g., diet, smoking, or medication use) cannot be taken into account in our study. 3) Due to the inherent constraints of MR, this study cannot elucidate the specific biological pathways linking IBD to chronic heart failure (CHF). Future studies combining multi-omics data and experimental models are needed. 4) The limited number of SNPs for CD (n = 4) and UC (n = 12) may reduce statistical power to detect smaller causal effects. 5) Potential sample overlap between exposure and outcome datasets, though minimized, cannot be entirely ruled out. The potential molecular mechanism can be explained and verified by the combination of bioinformatics analysis and experiments in the future.
In conclusion, the results of this study show that CD and UC will increase the risk of chronic heart failure at the genetic level of the European population. This finding is of great significance for in-depth understanding of the effects of IBD on chronic heart failure and the prevention and treatment of chronic heart failure. At the same time, we also need to pay attention to the limitations of the research results, and the relevant conclusions need to be further studied and verified.