Introduction
Diabetes is increasingly recognized as a significant global public health challenge. The worldwide diabetic population has escalated to 529 million in 2021 and is anticipated to rise to 1.31 billion by 2050 [1]. With the rising prevalence of diabetes mellitus and a trend towards younger patient populations, the global incidence of diabetic retinopathy (DR) has been on the rise. By 2045, it is anticipated that DR cases will increase to 160.5 million, impacting 44.82 million individuals with vision-threatening diabetic retinopathy (VTDR) [2]. DR, a significant microvascular complication, stands as a key contributor to vision impairment among patients. This condition stems from sustained damage to the retinal vasculature, resulting in alterations such as hard exudates, cotton-wool spots, and vascular remodeling [3].
Laser therapy, vitrectomy, anti-ceramide immunotherapy, and intravitreal injections represent available modalities for managing DR [4]. However, these interventions are predominantly aimed at slowing down DR progression rather than providing a definitive cure. Despite the existence of treatment options, the screening rate for DR remains below 50% due to various socio-environmental factors, including economic and regional disparities. Consequently, many patients who do not receive timely and regular treatment face irreversible visual impairment [5]. Therefore, the identification and management of DR risk factors are paramount.
There are numerous factors that influence the risk of developing DR [6]. Notably, lipid profiles within the diabetic cohort have garnered considerable global attention due to their distinct correlation with various medical conditions. Hyperlipidemia is a systemic metabolic disorder [7]. Dyslipidemia escalates the susceptibility to macrovascular complications such as peripheral vascular disease, coronary heart disease, and cerebrovascular disease, as well as microvascular issues such as retinopathy and end-stage renal disease in patients with diabetes [8, 9]. Research into the relationship between lipids and DR has evolved significantly over the past decades. Initial investigations have indicated that patients with DR exhibit elevated baseline lipid concentrations compared to the broader diabetic population [10]. A growing body of research suggests that this association may be due to the involvement of multiple lipid components. Consequently, early initiation of lipid-lowering medications not only reduces the incidence of other complications, but also significantly reduces mortality.
A diverse array of lipid-lowering medications are presently accessible for managing dyslipidemia. The evolution of therapeutic approaches has seen significant advancement, from the introduction of first-generation statins to the development of more targeted therapies. Guidelines, including those from the European Atherosclerosis Society (EAS), advocate for statins as the primary therapeutic option for addressing dyslipidemia in diabetic patients [11]. In addition, the combination of ezetimibe, PCSK9 inhibitors, and fibrates is frequently employed to augment lipid-lowering interventions [11, 12]. However, the evidence concerning the influence of commonly used lipid-lowering medications on the initiation and progression of DR remains contentious. Among the studies of fenofibrate drugs, the well-known Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) study and the Action to Control Cardiovascular Risk in Diabetes (ACCORD) study suggested that fenofibrate slows the progression of diabetic retinopathy. However, these studies also revealed differences in the drug’s efficacy in patients with different subtypes and severity of DR [13, 14]. The role of statins in DR management has been similarly debated over time. Early small-scale studies and extensive observational studies spanning the past two decades have indicated potential advantages of statins in mitigating late-stage complications of DR and averting vision impairment [15–17]. These findings seemed promising until a recent study suggested that statin usage might elevate the prevalence of DR in both proliferative and non-proliferative diabetic retinopathy (NPDR and PDR) subgroups, as well as in the broader DR population [18]. These conflicting outcomes underscore the critical need for additional comprehensive research to delineate the precise impact of lipid-lowering drugs on DR.
To overcome the limitations of observational studies, the use of MR methods leveraging comprehensive summary data from genome-wide association studies (GWAS) is gaining traction. MR harnesses genetic variations as inherent experiments to provide insights into potential causal relationships between risk factors and diseases, with reduced susceptibility to environmental confounders or reverse causation [19]. In drug target MR analysis, the simulation involves the pharmacological blockade of genetic drug targets utilizing pertinent genetic variants as instrumental variables, encompassing quantitative trait loci for expression (eQTLs) and protein (pQTLs). This approach is employed to assess the consequences of drug exposure [20]. Using a drug-target MR approach, this study simulates exposure to lipid-lowering drugs in diabetic patients to elucidate the causal relationship between these drugs and DR and to provide a basis for clinical strategies for the prevention and treatment of DR.
Study design
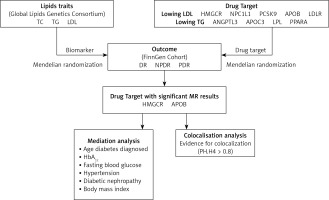
The recommendations for MR (STROBE-MR), which strengthens the reporting of observational studies in epidemiology, were followed in this investigation (Supplementary Table SI) [21]. Mendelian randomization (SMR) utilizing summary data and two-sample MR methods were employed to explore the relationship between diabetic retinopathy (DR) risk and targets of lipid-lowering medications. All data used in this investigation were sourced from published, publicly available summary statistics, as detailed in Supplementary Table SII. Approval from the respective ethics committees was obtained for all original studies. The study design workflow is depicted in Figure 1.
Data sources and selection of genetic instrumental variables
Lipid biomarkers
Genetic association data for lipid biomarkers were obtained from the Global Lipids Genetics Consortium, which represents the largest genome-wide association study (GWAS) meta-analysis to date, encompassing approximately 1.5 million people of European heritage [22]. The primary biomarkers considered were triglycerides (TG), total cholesterol (TC), and low-density lipoprotein cholesterol (LDL-C), demonstrating significance (p < 5 × 108). These biomarkers met the criteria of a physical distance requirement of 10,000 kb and a chain unbalance [LD] aggregation threshold of r2 < 0.001. To address potential sample overlap bias, particularly given that the outcome variables were derived from a Finnish database, participants from the Finnish Biobank (n = 177,987) were excluded from the dataset.
Lipid-lowering drug targets
Using information on both established and emerging lipid-lowering drugs [16, 23], we identified pertinent drug target genes using the DrugBank database. Subsequently, we conducted a comprehensive analysis integrating insights from existing literature and research findings [24, 25]. The study ultimately encompassed a total of 7 lipid-lowering drugs corresponding to 9 targets. Statins function by inhibiting 3-hydroxy-3-methylglutaryl-CoA reductase (HMG-CoA reductase), resulting in the upregulation of hepatic low-density lipoprotein receptors (LDL receptors). This mechanism enhances the efficiency of LDL clearance. On the other hand, ezetimibe operates by inhibiting the Niemann-Pick C1-Like 1 (NPC1L1) gene, which is accountable for cholesterol absorption in the intestine and liver, significantly reducing plasma total cholesterol and LDL-C levels. Proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors function by impeding the interaction of PCSK9 with LDL receptors (LDLR). This action prevents the degradation of LDL receptors by PCSK9, leading to an increase in the quantity of LDL receptors on the liver surface and a pronounced reduction in plasma LDL-C levels. Bile acid sequestrants operate by binding bile acids within the intestine, impeding their reabsorption and consequently lowering LDL-C levels. Mipomersen acts to diminish the synthesis of apolipoprotein B-100 (APOB-100), resulting in decreased levels of very low-density lipoprotein (VLDL), LDL, and lipoprotein(a) [9]. Fibrates specifically target peroxisome proliferator-activated receptor α (PPARα), enhancing the activity of lipoprotein lipase, thereby significantly reducing plasma triglyceride (TG) levels [26]. Angiopoietin-like 3 (ANGPTL3) functions by suppressing ANGPTL3 protein, thereby enhancing the activity of lipoprotein lipase. Antisense oligonucleotides directed at APOC3 mRNA serve to inhibit APOC3 synthesis, resulting in a notable decrease in plasma TG levels [27]. Additionally, lipoprotein lipase (LPL) mainly functions on the surface of capillary endothelial cells, catalyzing the hydrolysis of triglycerides within circulating triglyceride-rich lipoproteins such as chylomicrons and VLDL. Predicated on the fundamental pharmacological functions of these target genes, we subsequently categorized them into genes targeting the reduction of LDL-C and TG levels. Detailed information is summarized in Table I.
Table I
Characteristics of genetic instruments
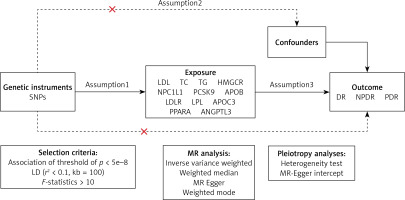
A systematic methodology was employed to ascertain pertinent genetic variants, drawing upon established methodologies from previous studies [28]. Our selection process involves two main steps. Firstly, we initially identified variants that achieved genome-wide significance (p < 5 × 10–8) within a 100 kb vicinity of the gene under investigation. This threshold is widely accepted in GWAS as indicative of strong evidence for an association. To ensure independence among the selected variants, we further refined our selection using LD-based clumping. We applied an LD threshold of r2 < 0.1. This step helps to mitigate the risk of including multiple correlated variants that could introduce biased estimates in subsequent analyses.
Genetic associations with diabetic retinopathy
Genetic association data for three outcomes were selected from the FinnGen R9 release GWAS summary statistics. The primary outcome was total DR, with secondary outcomes including NPDR and PDR. These outcomes were diagnosed according to the International Classification of Diseases, 10th Revision (ICD-10) classification system. Specifically, DR diagnosis primarily relied on the H36.0 code, with supplementary codes within the H35 category utilized for specific classifications. Specifically, H35.0 was used for NPDR diagnosis, while H35.2 was commonly used for PDR diagnosis. The sample sizes were as follows: DR included 10,413 cases and 308,633 controls; NPDR included 3,494 cases and 366,864 controls; PDR included 9,511 cases and 362,581 controls.
Statistical analysis
To investigate the causal relationship between cholesterol-lowering medications and genetically instrumented circulating lipid traits in relation to DR, NPDR, and PDR, we used two-sample MR analysis. To represent the impact of a 1 mmol/l shift in lipid levels, all estimates – given as odds ratios (ORs) – were normalized to be 38.7 mg/dl for LDL-C, 88.5 mg/dl for TG, and 38.7 mg/dl for TC.
For drug target genes showing positive associations with outcome variables in the two-sample analysis, we used the GTEx database to examine their expression in high-expression tissues. Subsequently, we conducted SMR analysis to evaluate the association between a 1-standard deviation (1-SD) change in drug target gene expression levels and outcome variables.
We utilized the Bonferroni adjustment for multiple testing, setting significance thresholds at p < 0.006 (0.05/9) for the nine pharmacological targets and p < 0.016 (0.05/3) for the three lipid characteristics. For all remaining analyses, statistical significance was defined as a two-sided p-value < 0.05. The statistical analyses were conducted using the R software (version 4.3.1) and involved the “TwoSampleMR”, “MendelianRandomization” and “coloc” packages.
The MR method is underpinned by three core assumptions [29]: exclusion limitation, independence, and relevance (Figure 2). Avoiding bias resulting from inadequate instrumental variables, we verified the strength of drug target instrumental variables by calculating F-statistics (β2/SE2), with F > 10 indicating sufficient instrument strength [30, 31]. Considering the well-documented advantages of lipid-lowering medication therapy in lowering the risk of coronary artery disease (CAD), we designated CAD as a positive control to confirm the efficacy of pertinent instrumental variables. Genetic association data for CAD were derived from a genome-wide association study involving 361,194 controls and 42,096 clinically diagnosed patients. To exclude bias from confounding factors beyond the study exposure, we conducted Bayesian colocalization analysis for drug targets significantly associated with outcome variables. Bayesian colocalization, founded on Bayes’ theorem, serves as a tool to evaluate whether distinct molecules (e.g., proteins, RNAs) are situated in close proximity in a cell, with the central idea being to combine prior knowledge with observed data to derive posterior probabilities of colocalization [32]. This analysis assessed whether drug targets and DR-related SNPs were driven by the same causal variant (posterior probability PP.H4) or influenced by different but linkage disequilibrium-related causal variants (PP.H3) [32]. A posterior probability exceeding 0.80 was considered indicative of support for the colocalization hypothesis.
To explore the specific pathways through which positive drug targets affect DR, we assessed the connection between recognized risk factors (e.g., age at diabetes diagnosis, fasting blood glucose, glycated hemoglobin (HbA1c), diabetic nephropathy, hypertension, body mass index (BMI)) for DR and genetically proxied lipid-lowering treatments [33]. Subsequently, after considering mediating effects, we evaluated these effects using a two-step MR approach. This approach enabled the quantification of the direct impact of genetically linked lipid-lowering medications on DR, the evaluation of the indirect influence of the mediator via the product of coefficients method, and the determination of the standard error of the indirect effect using the delta method. To confirm the strength and reliability of the findings, we conducted a test of heterogeneity (Cochran’s Q test) and a test of multiple validity (MR-Egger regression intercept test).
Results
Traits of lipids and DR risk
As instrumental factors for lipid characteristics, we found separate SNPs associated with TG, TC, and LDL-C (Supplementary Tables SIII–SV). Genetically proxied increases in TG levels were associated with a higher risk of DR in the general population, according to a two-sample MR analysis (OR = 1.34; 95% CI: 1.20–1.50; p = 1.25 × 10–7). However, no significant associations were observed for LDL-C and TC with DR or its subtypes in the overall population (Tables II and Supplementary Table SVII).
Table II
Risk association between blood lipids and DR
We performed genetic simulations for nine lipid-lowering drug targets (Supplementary Table SVIII). A positive control analysis was carried out to validate the effectiveness of the genetic instruments, revealing that eight genetically proxied pharmacological targets (excluding ANGPTL3) decreased CAD risk (Figure 2). The genetic instruments’ F-values varied from 10 to 5810, indicating that the potential influence of instrumental variable bias on the study outcomes was unlikely (Supplementary Table SVI).
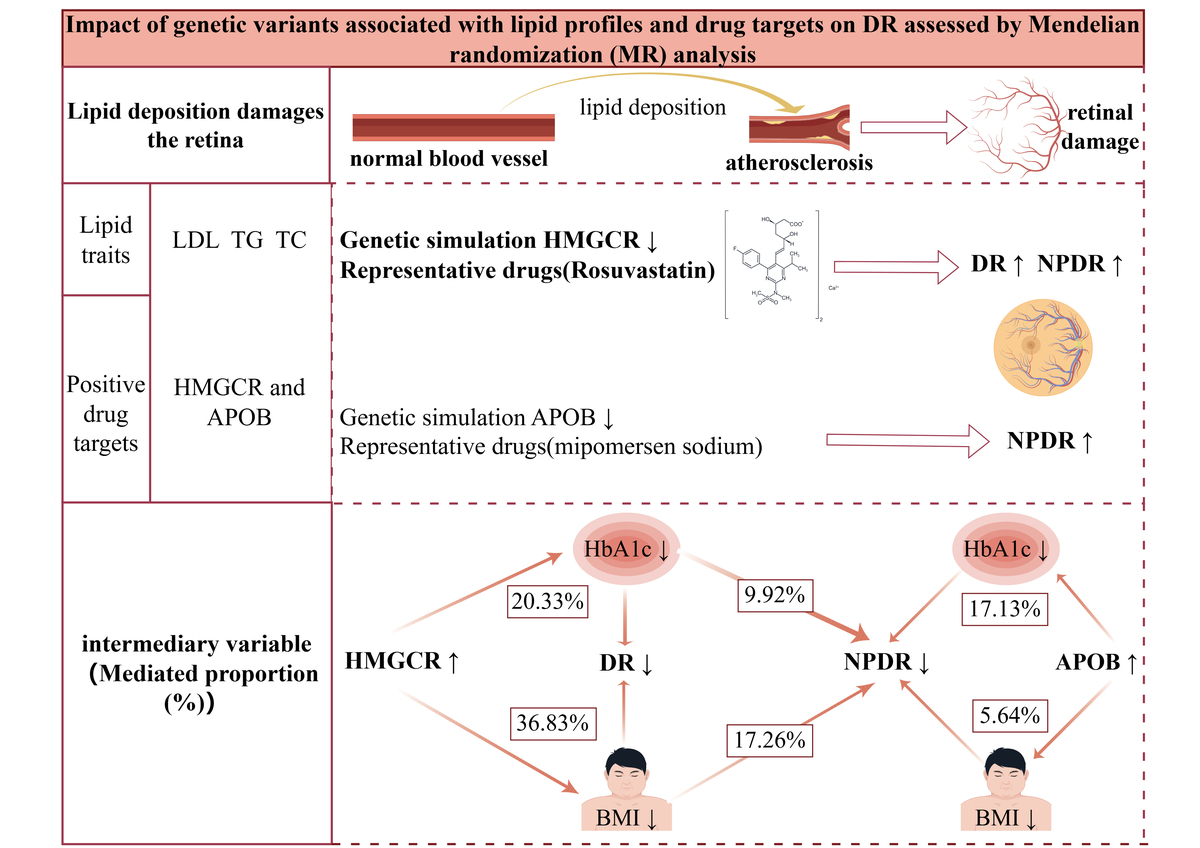
Figure 3 illustrates the effects of genetically proxied lipid-lowering drugs on DR. Our analysis revealed nuanced and differential effects across various drug targets. HMGCR targets showed the most consistent and significant protective effects with DR subtypes. In the DR (OR = 0.62; 95% CI: 0.46–0.83; p = 0.01) and NPDR (OR = 0.49; 95% CI: 0.34–0.70; p = 9.70 × 10–3) populations, genetically modelled HMGCR augmentation and a 1 mmol/l (88.9 mg/dl) rise in TG were associated with a lower risk of DR and NPDR. APOB targets showed consistent risk reduction across DR subtypes. Genetic simulation of APOB enhancement also showed associations with low DR risk (DR: OR = 0.75; 95% CI: 0.60–0.94; p = 0.01; NPDR: OR = 0.64; 95% CI: 0.48–0.87; p = 4.30 × 10–3; PDR: OR = 0.82; 95% CI: 0.69–0.99; p = 0.03). Notably, the protective effect was most pronounced in the NPDR subgroup. Conversely, genetic simulation of ANGPTL3 enhancement was observed to increase the risk of DR and PDR. However, the significance of the associations of ANGPTL3 and APOB with outcomes diminished after Bonferroni correction, warranting cautious interpretation. Notably, no significant relationships were identified between the other genetically simulated drug targets and DR outcomes.
The outcomes obtained from alternative analysis approaches were largely consistent with those from the primary analysis method (inverse variance-weighted method) (Supplementary Table SVIII). No indications of pleiotropy were detected for the variables, except in the analyses involving LDL-C and TG, where the MR-Egger intercepts exceeded 0, thereby enhancing the validity of causal inference.
Gene expression and DR risk
Blood, liver, and subcutaneous adipose tissues exhibiting the highest expression levels of HMGCR, APOB, and ANGPTL3 genes were selected for SMR analysis using the GTEx database. The results revealed a significant correlation between a 1-standard deviation (1-SD) rise in HMGCR expression in blood tissue with a lower incidence of DR (DR: OR = 0.66; 95% CI: 0.52–0.85; p = 7.31 × 10–4; NPDR: OR = 0.64; 95% CI: 0.44–0.93; p = 2.03 × 10–2) (Supplementary Table SX). No significant associations were identified between genes related to APOB or ANGPTL3 and DR or NPDR. Colocalization analysis was conducted to determine the likelihood of shared causal SNPs between genetic variants associated with HMGCR expression in whole blood tissue and DR/NPDR. The results revealed a common causal variant for HMGCR expression in whole blood tissue and DR (PP.H4 = 0.85) (Supplementary Table SXI), but no strong evidence for a shared causal variant with NPDR (PP.H4 = 0.27).
Mediation analysis
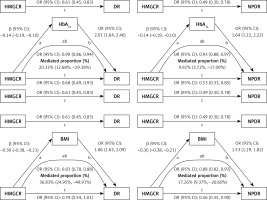
To explore mediating factors in HMGCR’s influence on DR risk, we performed a two-step MR analysis of six potential mediating variables to assess their role in the effects of HMGCR and APOB on DR. The results showed significant causal associations between HMGCR and HbA1c, BMI, and hypertension, whereas APOB was causally associated with HbA1c and BMI (Supplementary Table SXII). For HMGCR, we found that HbA1c mediated 9.92% (95% CI: 3.72%, 17.09%) of the total effect of HMGCR on NPDR and 20.33% (95% CI: 12.68%, 29.38%) of the total effect of HMGCR on DR. The mediating effect of BMI was more significant, accounting for 17.26% (95% CI: 9.37%, 26.60%) of the total effect of HMGCR on NPDR and 36.83% (95% CI: 24.95%, 49.91%) of the total effect on DR (Figure 4). The mediating effect of hypertension, although statistically significant, was relatively small, accounting for only 2.34% (95% CI: 0.14%, 5.65%) of the total effect of HMGCR on NPDR and 3.82% (95% CI: 0.40%, 8.25%) of the total effect on DR. For APOB, the mediating effect of HbA1c in its total effect on NPDR was 11.73% (95% CI: 4.06%, 21.21%), while the mediating effect of BMI was 5.64% (95% CI: 2.48%, 9.70%). These results suggest that the genetically modelled effects of HMGCR and APOB on reducing the risk of DR are partly mediated by these mediators. In particular, BMI and HbA1c played a large mediating role in the effect of HMGCR on DR, whereas the mediating role of HbA1c was more significant in the effect of APOB on NPDR. Detailed statistical results are presented in Supplementary Tables SXII–SXIV.
Discussion
This study examined the possible effects of lipid-lowering treatment targets while using MR and drug target SMR methodologies to explore the causal link between blood lipid levels and DR [34]. The key findings of our study are as follows: elevated TG levels significantly increase the risk of DR and NPDR; HMGCR is negatively correlated with DR risk in the total population and NPDR; APOB is also negatively correlated with NPDR risk. Nevertheless, there is no proof that lipid characteristics or the nine pharmacological lipid-lowering targets have an effect on PDR. Mediation analyses highlight the importance of glycemic control and weight management in the prevention and management of DR. Although hypertension is a known risk factor for DR, the role of HMGCR-related lipid-lowering drugs in DR appears to be less prominent than that of BMI and HbA1c.
The correlation between DR risk and blood lipid levels remains controversial. Previous studies have reported conflicting results, with some investigations showing no clear relationship between any blood lipid component and any form of DR [35, 36]. For instance, a MR study conducted by Sobrin et al. did not identify any causal effect of four lipid components (HDL, LDL, TG, TC) on DR [33]. However, a study by Dornan et al. suggested that LDL levels were higher in PDR populations compared to NPDR and normal populations, implying a correlation between LDL and DR severity [37]. Furthermore, a large Spanish follow-up study reported that LDL levels and the TC to LDL ratio were associated with an increased risk of DR [34, 38]. These findings align with a meta-analysis of 13 cohort studies, which indicated that baseline TG levels were associated with the development of diabetic retinopathy in individuals with diabetes, while no significant associations were found between the prevalence of diabetic retinopathy and LDL and TC levels [39]. This inconsistency reflects the complex relationship between lipids and DR, possibly related to methodological differences, population characteristics, or different stages of DR.
In the current study, genetic simulation of HMGCR and APOB was associated with a decreased risk of any DR and NPDR, which is consistent with the findings of Chen et al. However, this protective effect does not seem to extend to PDR. Additionally, mediation analyses demonstrated that the protective impact of HMGCR on DR was partially mediated by HbA1c and BMI, with the causal effect of HMGCR on these two mediating variables resembling previous findings [40, 41]. An animal model-based investigation revealed that reduced HMGCR expression led to elevated dietary intake and fat storage, potentially mediated by target of brain insulin (TOBI) regulation through modulation of α-glucosidase gene expression to regulate blood glucose levels [42]. This discovery implies that genetic mimicry of HMGCR to promote blood glucose and BMI reduction may be associated with this phenomenon. The study further confirmed the association between HMGCR and BMI by observing weight gain resulting from statin drug administration [43]. Chronic hyperglycemia induces oxidative stress by increasing reactive oxygen species (ROS) production, triggering an inflammatory cascade resulting in vascular wall damage and increased vascular permeability, activation of protein kinase C and advanced glycation end-products (AGEs), and endothelial dysfunction. In addition, sustained high blood glucose levels lead to pericyte detachment and basement membrane thickening, impaired neurovascular coupling, and disruption of retinal microcirculation. These series of metabolic and physiological disturbances ultimately induce the development of DR.
Additionally, genetic mimicry of APOB associated with lowering LDL-C was identified as contributing to the reduction in NPDR risk. It is noteworthy that the impact of apolipoprotein B (APOB) on DR in previous investigations was multifaceted, with some studies indicating that APOB increased retinal small artery tortuosity, a factor correlated with the severity of DR [44, 45]. However, Li et al. did not find an association between genetically determined APOB and DR risk. This inconsistency may prompt us to revisit the relationship between APOB and DR. It is possible that genetic variations could influence the function of APOB beyond its levels, and specific instrumental variables related to the APOB gene might exert protective effects. Furthermore, the complex interplay of genetic background, environmental factors, and population variances may all contribute to the diverse outcomes observed in this association.
Previous large clinical studies such as FIELD and ACCORD-EYE have demonstrated the protective effect of betablockers in patients with diabetic retinopathy (DR) [46]. However, in the present study, only inhibition of the ANGPTL3 gene showed a suggestive, but not significant, association with reduced risk of DR. This discrepancy may stem from several aspects: first, fibrates such as fenofibrate exert their triglyceride (TG) lowering effects by activating multiple signaling pathways, rather than relying on a single pathway. This multi-targeted action may be an important reason for the superior therapeutic effect over single gene inhibition observed in clinical trials. Second, although studies have confirmed the correlation between certain circulating lipid levels and DR progression, the final clinical phenotype is modulated by multiple factors. In addition to changes at the transcriptional and translational levels of genes, environmental factors, epigenetic modifications, and other unknown regulatory mechanisms may influence disease progression and therapeutic efficacy. This complex interaction may lead to differences in the effects of single gene interventions versus the results of drug therapy.
These seemingly contradictory findings suggest that lipid-lowering drugs may have different mechanisms and effects in the prevention and treatment of DR. It was found that microglia aggregation and systemic inflammation were more severe in patients with PDR, whereas fenofibrate ameliorated oxidative stress and systemic inflammation, while also inhibiting infiltration and activation of retinal cells [47]. Previous studies have shown that the degree of systemic inflammation in patients with DR is related to the grade of the disease, and the differences in the efficacy of different types of lipid-lowering drugs in patients with different DR grades may be related to the above factors. Therefore, it is important to explore the mechanism of action of these drugs in different types of DR, especially the relationship with inflammatory reactions and cell activation.
The pathological process of DR is complex, with evidence indicating that plasma LDL-C and cholesterol levels are associated with retinal hard exudates [48, 49]. Statin medications have been shown by Gupta et al. to lessen the intensity of hard exudates and central foveal lipid migration [18, 50], further supporting the association between blood lipids and retinal exudation. The intricate relationship between circulating lipids and DR has been confounded by numerous factors, prompting an increasing number of studies to explore and emphasize non-lipid mechanisms. Recent research has indicated that disruption of retinal cholesterol metabolism and impaired retinal capillary repair may serve as the underlying mechanisms of DR. Initially, retinal cholesterol accumulation leads to the formation of highly reflective crystalline deposits (CCS), which activate the immune response, triggering the NLRP3 inflammasome and the release of various inflammatory factors, such as IL-1, thereby inciting local tissue inflammation [47]. Various microbial infections activate Toll-like receptors (TLRs), inhibit liver X receptors (LXRs) through the viral response to the transcription factor interferon regulatory factor 3 (IRF3), reduce expression of ABCA1 transporter proteins, and inhibit cholesterol efflux by macrophages [51]. In addition, chronic inflammatory activation in diabetic patients can disrupt bone marrow microenvironmental homeostasis and slow retinal vascular endothelial cell repair, thus exacerbating the progression of DR [52].
This study leveraged the advantages of large-scale samples from the Finnish Biobank, providing important insights into the genetic risk of DR. It demonstrated the potential for more targeted and individualized interventions beyond the current “one-size-fits-all” approach. The research methods employed helped to avoid the reverse causality issues and confounding variables that are prevalent in traditional observational studies. From a clinical application perspective, our findings provide new opportunities for precision medicine. It is worth noting that although the study found that genetic mimicry of HMGCR and APOB enhancement were beneficial in reducing the risk of DR, we acknowledge the therapeutic benefits of lipid-lowering medications such as statins and mipomersen in individuals with diabetes. Our findings offer a nuanced approach to clinical implementation for patients with DR and hyperlipidemia. A comprehensive assessment of the patient’s genetic and metabolic profile is recommended to develop a personalized lipid-lowering strategy for optimal management based on the patient’s individual profile. Future research endeavors may focus on identifying the most suitable lipid-lowering approach based on the unique characteristics of patients (e.g., DR stage, genotype).
However, we also acknowledge the limitations of the methodology of this study: (1) Despite conducting sensitivity analyses, studies based on GWAS data still cannot completely rule out pleiotropy, there may be other confounding factors affecting outcomes, and the results have the potential to be false-positive. (2) Given that the GWAS data originated from European cohorts, caution is warranted in generalizing the findings to other ethnic populations, as population-specific effects may not be accurately represented. (3) Although the study detailed specific retinal effects of lipid-lowering drugs in patients with DR, the GWAS-based data were limited, and it was not possible to obtain the eQTL data of HMGCR and APOB in retinal tissues or the results for the intraretinal effects. (4) The potential disparities between the direct effects of lipid-lowering therapies and the effects of genetic variants on DR risk were not directly evaluated in this study. (5) The original GWAS data can only be used to make the main classification based on DR, but the progression of DR includes many other important pathological processes, and refinement of the effects of lipid-lowering therapies on these pathological processes will help to strengthen the results, thereby facilitating a more comprehensive interpretation of the results.
We acknowledge several critical limitations in our current study that necessitate future research. Firstly, the multiplicity of studies can be reduced by developing advanced statistical methods to more accurately distinguish between direct and indirect genetic effects and by using a multi-omics approach to reveal complex genetic interactions. Secondly, in order to generalize the results of the study, multi-ethnic studies can be conducted at a later stage to incorporate different genetic backgrounds to ensure the robustness of the risk assessment. Stratified analysis protocols could also be developed to take into account genetic variation in specific populations.
Based on our findings, future research directions may include: 1) further elucidating the mechanisms of action of lipid-lowering drugs at different DR stages, especially non-lipid effects; 2) developing comprehensive genetic screening protocols, exploring individualized treatment strategies based on genotype risk prediction and treatment selection, and developing predictive models that integrate genetic, metabolic, and clinical variables; 3) creating algorithm-based treatment selection models, considering the interaction between DR and other diabetic complications and developing an integrated management approach that considers multiple metabolic pathways; 4) conducting longitudinal studies investigating long-term outcomes of lipid management in DR populations, ongoing monitoring of DR progression, and validation of HMGCR and APOB-targeted therapies. This will lead to a comprehensive assessment of potential side effects and long-term efficacy; 5) combining genetic insights with clinical practice for personalized diabetic retinopathy management and developing precision medicine approaches.
In conclusion, our research used MR and SMR techniques to illuminate the intricate connection between lipid metabolism and DR, particularly the potential protective role of HMGCR and APOB in NPDR risk. The mediation analysis highlighted the importance of glycemic control and weight management in the prevention and management of DR. These results contribute to our understanding of the pathogenic underpinnings of DR and offer fresh perspectives for preventative and therapeutic approaches in the future. While further research is required to confirm and elaborate on these findings, our study translates genetic insights into practical clinical applications, ultimately improving patient outcomes and developing more precise, personalized approaches to diabetic retinopathy management.