Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
NEPHROLOGY / RESEARCH PAPER
Metabolic profiles of IgA nephropathy, membranous nephropathy, and diabetic nephropathy
1
Department of Nephrology, Fifth Hospital of Shanxi Medical University (Shanxi Pronvincial People’s Hosipital), Taiyuan, China
2
Department of Medicine, International College of Medicine and Pharmacy, Changsha Medical University, Changsha, China
3
Key Laboratory of Kidney Disease, Precision Medicine Center, Shanxi Provincial People's Hospital, Shanxi Medical University, Taiyuan, China, China
Submission date: 2025-02-12
Final revision date: 2025-08-10
Acceptance date: 2025-09-12
Online publication date: 2026-02-08
Corresponding author
Rongshan Li
1.Department of Nephrology, Fifth Hospital of Shanxi Medical University (Shanxi Pronvincial People’s Hosipital)Taiyuan, China, China
1.Department of Nephrology, Fifth Hospital of Shanxi Medical University (Shanxi Pronvincial People’s Hosipital)Taiyuan, China, China
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Immunoglobulin A nephropathy (IgAN), membranous nephropathy (MN), and diabetic nephropathy (DN) are prominent contributors to chronic kidney disease burden. Our main objective was to contribute to understanding of metabolic profiles of these three major types of nephropathies and identify potential metabolic biomarkers.
Material and methods:
Kidney samples of 20 sex- and age-matched patients with biopsy-proven IgAN, MN, DN, and controls without any kidney diseases were included. Ultra high performance liquid chromatography-mass spectrometry analysis was conducted. t-test was used to calculate statistical significance of the identified metabolites. Metabolic pathways were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG). Specificity, sensitivity and area under the curve (AUC) were calculated to evaluate the predictive performance of metabolites.
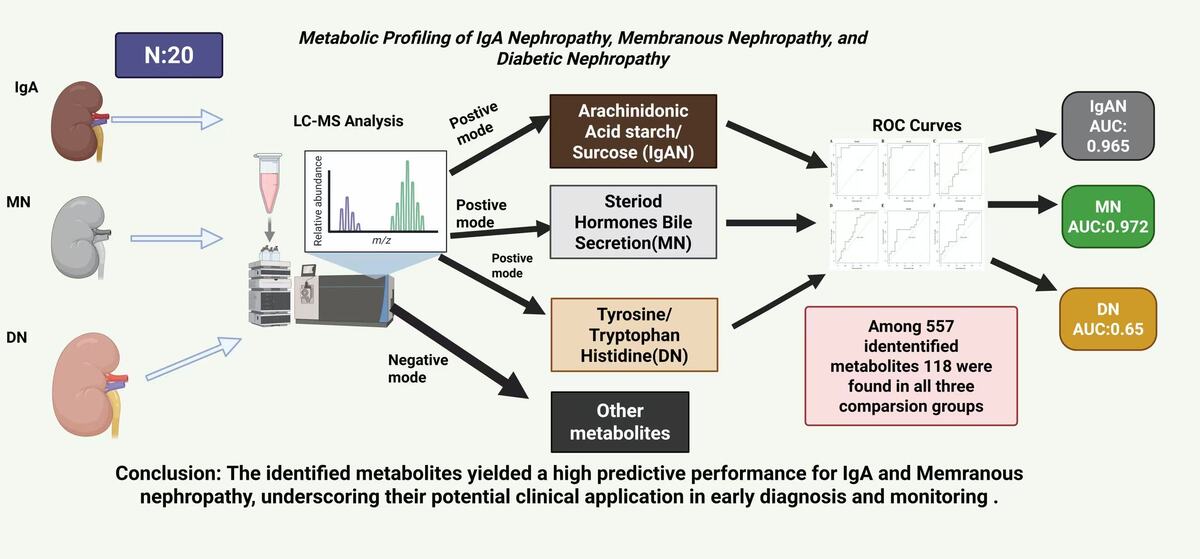
Results:
Among 557 identified differential metabolites, only 118 were found in all three comparison groups. Differential metabolites of IgAN vs controls were significantly enriched in arachidonic acid metabolism, starch and sucrose metabolism, ferroptosis, and other pathways. In the DN group, metabolites were mainly enriched in phenylalanine, tyrosine and tryptophan biosynthesis, histidine metabolism, etc. MN-enriched pathways included steroid hormone biosynthesis, neuroactive ligand-receptor interaction, and bile secretion. In the positive mode, cumulative AUC values for comparison pairs IgAN vs controls, MN vs controls, and DN vs controls were 0.965, 0.972, and 0.573, respectively, whereas in the negative mode the AUC values of all three pairs were slightly above 0.65.
Conclusions:
IgAN, MN and DN have similar but distinct metabolic profiles. Only positive node metabolites of IgAN and MN exhibited great predictive performance.
Immunoglobulin A nephropathy (IgAN), membranous nephropathy (MN), and diabetic nephropathy (DN) are prominent contributors to chronic kidney disease burden. Our main objective was to contribute to understanding of metabolic profiles of these three major types of nephropathies and identify potential metabolic biomarkers.
Material and methods:
Kidney samples of 20 sex- and age-matched patients with biopsy-proven IgAN, MN, DN, and controls without any kidney diseases were included. Ultra high performance liquid chromatography-mass spectrometry analysis was conducted. t-test was used to calculate statistical significance of the identified metabolites. Metabolic pathways were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG). Specificity, sensitivity and area under the curve (AUC) were calculated to evaluate the predictive performance of metabolites.
Results:
Among 557 identified differential metabolites, only 118 were found in all three comparison groups. Differential metabolites of IgAN vs controls were significantly enriched in arachidonic acid metabolism, starch and sucrose metabolism, ferroptosis, and other pathways. In the DN group, metabolites were mainly enriched in phenylalanine, tyrosine and tryptophan biosynthesis, histidine metabolism, etc. MN-enriched pathways included steroid hormone biosynthesis, neuroactive ligand-receptor interaction, and bile secretion. In the positive mode, cumulative AUC values for comparison pairs IgAN vs controls, MN vs controls, and DN vs controls were 0.965, 0.972, and 0.573, respectively, whereas in the negative mode the AUC values of all three pairs were slightly above 0.65.
Conclusions:
IgAN, MN and DN have similar but distinct metabolic profiles. Only positive node metabolites of IgAN and MN exhibited great predictive performance.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.